| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,836,162 – 14,836,258 |
| Length | 96 |
| Max. P | 0.977171 |

| Location | 14,836,162 – 14,836,258 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.97 |
| Mean single sequence MFE | -20.46 |
| Consensus MFE | -10.65 |
| Energy contribution | -11.32 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
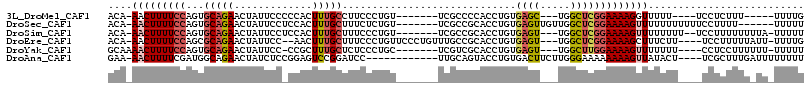
>3L_DroMel_CAF1 14836162 96 + 23771897 ACA-AACUUUUCCAGUGCAGAACUAUUCCCCCACUUUGCCUUCCCUGU-------UCGCCCCACCUGUGAGC---UGGCUCGGAAAAGGUUUUU----UCCUCUUU-----UUUUG ...-..(((((((((.(((((.............)))))))..((.((-------((((.......))))))---.))...)))))))......----........-----..... ( -20.22) >DroSec_CAF1 12863 102 + 1 ACA-AACUUUUCCAGUGCAGAACUAUUCCUCCACUUUGCUUUCUCUGU-------UCGCCGCACCUGUGAGUUGUUGGCUCGGAAAAGUUUUUUUUUUUCCUUUU------UUUUU ..(-(((((((((.((((.((((.......................))-------))...))))....((((.....))))))))))))))..............------..... ( -22.70) >DroSim_CAF1 13419 102 + 1 ACA-AACUUUUCCAGUGCAGAACUAUUCCUCCACUUUGCUUUCCCUGU-------UCGCCGCACCUGUGAGU---UGGCUCGGAAAAGUUUUUUUU--UCCUUUUUUUUA-UUUUU ..(-(((((((((.((((.((((.......................))-------))...))))....(((.---...))))))))))))).....--............-..... ( -21.80) >DroEre_CAF1 14810 105 + 1 ACA-AACUUUUCCAGCGCAGAACUAUUCC--AACUUUGCUUUCCCUGUUCCCUGUUUGCCGCACCUGUGAGU---UGGCUCGGAAAAGCUUUCUU----UCCUUUUUAUU-UUUUG ...-..(((((((((((((((........--...)))))..................(((((....))).))---..))).))))))).......----...........-..... ( -17.00) >DroYak_CAF1 8976 100 + 1 GCAAAACUUUUCCAGUGCAGAACUAUUCC-CCGCUUUGCUCUCCCUGC-------UCGUCGCACCUGUGAGU---UGGCUUGGAAAAGUUUUUUU----CCUCCUUUUUU-UUUUU ..(((((((((((((.(((((.(......-..).))))).)..((.((-------(((.((....)))))))---.))..))))))))))))...----...........-..... ( -23.10) >DroAna_CAF1 7848 99 + 1 GAA-AACUUUUCGAUGGCAGAACUAUCUCCGGAGUCCGGAUCC------------UUGCAGUACCUGUGACUUCUUGGGAAAAAAAAGUUAUACU----UCGCUUUGAUUUUUUUU ...-...((..(((.((.(((....)))))(((((((((....------------.........))).)))))))))..))((((((((((....----......)))))))))). ( -17.92) >consensus ACA_AACUUUUCCAGUGCAGAACUAUUCC_CCACUUUGCUUUCCCUGU_______UCGCCGCACCUGUGAGU___UGGCUCGGAAAAGUUUUUUU____CCUCUUUU_UU_UUUUU ....(((((((((...(((((.............))))).............................((((.....))))))))))))).......................... (-10.65 = -11.32 + 0.67)
| Location | 14,836,162 – 14,836,258 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.97 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | -12.17 |
| Energy contribution | -13.48 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
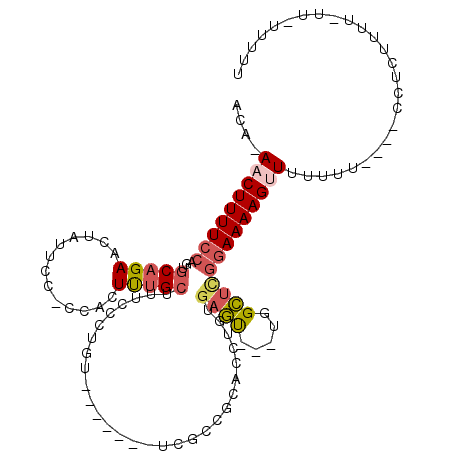
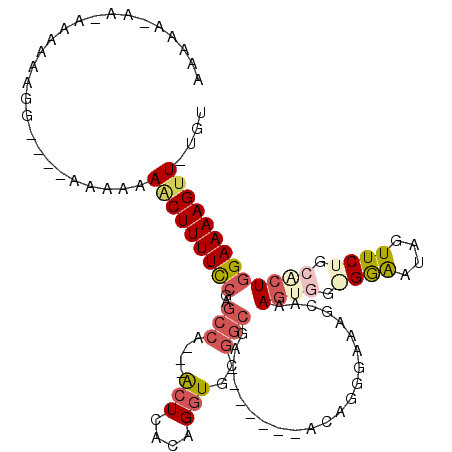
>3L_DroMel_CAF1 14836162 96 - 23771897 CAAAA-----AAAGAGGA----AAAAACCUUUUCCGAGCCA---GCUCACAGGUGGGGCGA-------ACAGGGAAGGCAAAGUGGGGGAAUAGUUCUGCACUGGAAAAGUU-UGU .....-----........----......(((((((..(((.---.(((....((.......-------)).)))..)))..((((.((((....)))).)))))))))))..-... ( -23.30) >DroSec_CAF1 12863 102 - 1 AAAAA------AAAAGGAAAAAAAAAAACUUUUCCGAGCCAACAACUCACAGGUGCGGCGA-------ACAGAGAAAGCAAAGUGGAGGAAUAGUUCUGCACUGGAAAAGUU-UGU .....------..............((((((((((..(((....(((....)))..)))..-------.............((((.((((....)))).)))))))))))))-).. ( -24.50) >DroSim_CAF1 13419 102 - 1 AAAAA-UAAAAAAAAGGA--AAAAAAAACUUUUCCGAGCCA---ACUCACAGGUGCGGCGA-------ACAGGGAAAGCAAAGUGGAGGAAUAGUUCUGCACUGGAAAAGUU-UGU .....-............--.....((((((((((..(((.---(((....)))..)))..-------.............((((.((((....)))).)))))))))))))-).. ( -25.70) >DroEre_CAF1 14810 105 - 1 CAAAA-AAUAAAAAGGA----AAGAAAGCUUUUCCGAGCCA---ACUCACAGGUGCGGCAAACAGGGAACAGGGAAAGCAAAGUU--GGAAUAGUUCUGCGCUGGAAAAGUU-UGU .....-...........----....(((((((((((((...---.)))...((((((.....)..(((((......(((...)))--......)))))))))))))))))))-).. ( -23.20) >DroYak_CAF1 8976 100 - 1 AAAAA-AAAAAAGGAGG----AAAAAAACUUUUCCAAGCCA---ACUCACAGGUGCGACGA-------GCAGGGAGAGCAAAGCGG-GGAAUAGUUCUGCACUGGAAAAGUUUUGC .....-...........----...((((((((((((.((..---.(((.(...(((.....-------))).)))).))...((((-((.....))))))..)))))))))))).. ( -25.70) >DroAna_CAF1 7848 99 - 1 AAAAAAAAUCAAAGCGA----AGUAUAACUUUUUUUUCCCAAGAAGUCACAGGUACUGCAA------------GGAUCCGGACUCCGGAGAUAGUUCUGCCAUCGAAAAGUU-UUC ....(((((.....(((----.((...(((((((......))))))).)).((((..((..------------...(((((...)))))....))..)))).)))....)))-)). ( -17.80) >consensus AAAAA_AA_AAAAAAGG____AAAAAAACUUUUCCGAGCCA___ACUCACAGGUGCGGCGA_______ACAGGGAAAGCAAAGUGG_GGAAUAGUUCUGCACUGGAAAAGUU_UGU ..........................(((((((((..(((....(((....)))..)))......................((((.((((....)))).))))))))))))).... (-12.17 = -13.48 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:49 2006