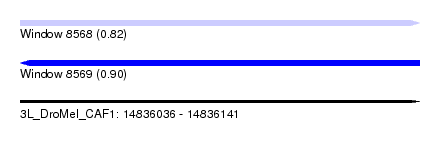
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,836,036 – 14,836,141 |
| Length | 105 |
| Max. P | 0.902316 |

| Location | 14,836,036 – 14,836,141 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 76.29 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -15.02 |
| Energy contribution | -15.00 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14836036 105 + 23771897 GAACG---GAGGAUAUUGAAUUCAUCGUACAAUAUGGCAAAGUCAAUAACAUGACCAUGUACGAGACAGUUGCCAAAUUGGAAAAUGUGAGUAUAUA--------U--CGGUGUUGGA .....---...((((((((((((((....((((.((((((.(((....((((....))))....)))..)))))).))))......)))))).....--------)--)))))))... ( -24.20) >DroVir_CAF1 35162 110 + 1 AUGCUAAUGAUCAUAUUGAAUUCGUUGUUCAAUAUGGCCGAGUCAAUAACAUGACCAUGUACGAGACGAUUGCCAAAUUGGAAAACGUGAGUACAGCUAAACAUAG--UAGU------ .(((((..(..((((((((((.....))))))))))..)(.((((......))))).(((((...(((....((.....))....)))..)))))........)))--))..------ ( -27.40) >DroPse_CAF1 68801 104 + 1 AAGCG---GAGGAUAUUGAUUUUAUUGUGCAGUAUGGCAGAGUCAAUAACAUGACCAUGUACGAGACAGUUGCCAAAUUGGAAAAUGUGAGUAUAUU--------U--CUGG-AUGGA ...((---(((((((((.((.......(.((((.((((((.(((....((((....))))....)))..)))))).)))).)....)).))))).))--------)--))).-..... ( -23.60) >DroGri_CAF1 32200 105 + 1 AAGCUAAUGAUAGAAUUGAAUUUGUCGUUCAAUAUGGCCGAGUCAAUAACAUGACCAUGUACGAGACAGUUGCCAAAUUGGAAUAUGUGAGUACAUC-----AUAU--GUGU------ .......(((((((......)))))))((((((.((((...(((....((((....))))....)))....)))).))))))((((((((.....))-----))))--))..------ ( -24.20) >DroWil_CAF1 160098 105 + 1 CUGCU---GAUGAUAUUGAUUUUAUUGUACAAUACGGCAAAGUCAAUGACAUGACCAUGUAUGAGACAGUUGCCAAAUUAGAAAAUGUAAGUAUCCA--------AGUUAG--CAAUA .((((---(((((((((.(((((...(((...)))(((((.(((....((((....))))....)))..))))).......)))))...))))))..--------.)))))--))... ( -23.60) >DroPer_CAF1 66258 104 + 1 AAGCG---GAGGAUAUUGAUUUUAUUGUGCAGUAUGGCAGAGUCAAUAACAUGACCAUGUACGAGACAGUUGCCAAAUUGGAAAAUGUGAGUAUAUU--------U--CUGG-AUGGA ...((---(((((((((.((.......(.((((.((((((.(((....((((....))))....)))..)))))).)))).)....)).))))).))--------)--))).-..... ( -23.60) >consensus AAGCG___GAGGAUAUUGAAUUUAUUGUACAAUAUGGCAGAGUCAAUAACAUGACCAUGUACGAGACAGUUGCCAAAUUGGAAAAUGUGAGUAUAUA________U__CUGU__UGGA ......................((((...((((.((((((.(((....((((....))))....)))..)))))).))))...))))............................... (-15.02 = -15.00 + -0.02)

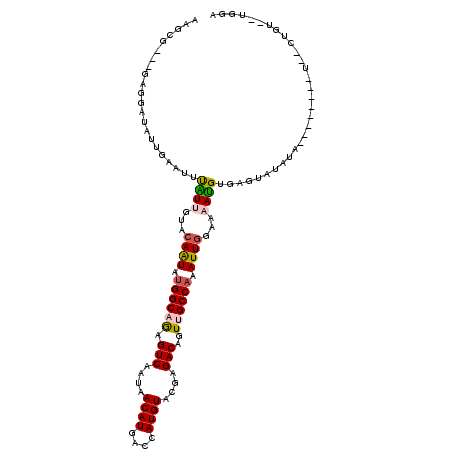

| Location | 14,836,036 – 14,836,141 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 76.29 |
| Mean single sequence MFE | -18.14 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14836036 105 - 23771897 UCCAACACCG--A--------UAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUUUGCCAUAUUGUACGAUGAAUUCAAUAUCCUC---CGUUC .........(--(--------(((.......((((...((((.((((((..(((....((((....))))....))).)))))).))))...))))......)))))...---..... ( -21.12) >DroVir_CAF1 35162 110 - 1 ------ACUA--CUAUGUUUAGCUGUACUCACGUUUUCCAAUUUGGCAAUCGUCUCGUACAUGGUCAUGUUAUUGACUCGGCCAUAUUGAACAACGAAUUCAAUAUGAUCAUUAGCAU ------....--.........(((((((..(((.((.((.....)).)).)))...))))..(((((......))))).(..(((((((((.......)))))))))..)...))).. ( -24.10) >DroPse_CAF1 68801 104 - 1 UCCAU-CCAG--A--------AAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUCUGCCAUACUGCACAAUAAAAUCAAUAUCCUC---CGCUU .....-....--.--------......................(((((...(((....((((....))))....)))..)))))..........................---..... ( -12.90) >DroGri_CAF1 32200 105 - 1 ------ACAC--AUAU-----GAUGUACUCACAUAUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUCGGCCAUAUUGAACGACAAAUUCAAUUCUAUCAUUAGCUU ------....--..((-----((((.............((((.((((....(((....((((....))))....)))...)))).))))...((.....)).....))))))...... ( -18.00) >DroWil_CAF1 160098 105 - 1 UAUUG--CUAACU--------UGGAUACUUACAUUUUCUAAUUUGGCAACUGUCUCAUACAUGGUCAUGUCAUUGACUUUGCCGUAUUGUACAAUAAAAUCAAUAUCAUC---AGCAG ..(((--((((.(--------((((...........))))).)))))))((((......((.(((((......))))).))..((((((...........))))))....---.)))) ( -19.80) >DroPer_CAF1 66258 104 - 1 UCCAU-CCAG--A--------AAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUCUGCCAUACUGCACAAUAAAAUCAAUAUCCUC---CGCUU .....-....--.--------......................(((((...(((....((((....))))....)))..)))))..........................---..... ( -12.90) >consensus UCCA__ACAG__A________AAUAUACUCACAUUUUCCAAUUUGGCAACUGUCUCGUACAUGGUCAUGUUAUUGACUCUGCCAUAUUGAACAAUAAAAUCAAUAUCAUC___AGCUU ......................................((((.(((((...(((....((((....))))....)))..))))).))))............................. (-13.69 = -14.08 + 0.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:47 2006