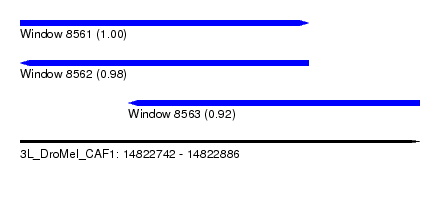
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,822,742 – 14,822,886 |
| Length | 144 |
| Max. P | 0.998085 |

| Location | 14,822,742 – 14,822,846 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -22.04 |
| Energy contribution | -25.44 |
| Covariance contribution | 3.40 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14822742 104 + 23771897 AAAGGCAAAAGGCAAGUGCAACUGCGUUUGCCACUGCAG--------------GUUGCAUAUUGCUCUGCUAUGCUCUUUACAACUUGUAUCAAGUUGU--UUUGGUGCUUUCUAGUUCU ...((((...((((((((((((((((........)))).--------------))))))).))))).))))..((.((..((((((((...))))))))--...)).))........... ( -32.80) >DroSec_CAF1 57533 104 + 1 AAAAGCAAAAGGCAAGUGCAACUGCGUUUGCCACUGCAG--------------GUUGCAGAUUGCUCUGCUAUGCUCUUUACAACUUGCAUCAAGUUGU--UUUGGUGCUUUCUAGCUCU ...(((((..(((((((((....))))))))).((((..--------------...)))).)))))..((((.((.((..((((((((...))))))))--...)).))....))))... ( -36.70) >DroSim_CAF1 50733 104 + 1 AAAGGCAAAAGGCAAGUGCAACUGCGUUUGCCACUGCAG--------------GUUGCAGAUUGCUCUGCUAUGCUCUUUACAACUUGCAUCAAGUUGU--UUUGGUGCUUUCUAGCUCU ..((((((..(((((((((....))))))))).((((..--------------...)))).)))).))((((.((.((..((((((((...))))))))--...)).))....))))... ( -36.80) >DroYak_CAF1 60844 118 + 1 AAAGGCAAAAGGCAAGUGCAACUGCGUUUGCCACUGCAGGUUGCUGGUUGCAGGUUGCAGAUUGCUCUGCAAUGCUCUUUACAACUUCCAUCAAGUUGU--UUUGGUGUUUUUUAGCUCU ..((((((..(((((((((....))))))))).((((((.((((.....)))).)))))).)))).))((((.((.((..(((((((.....)))))))--...)).))...)).))... ( -39.80) >DroAna_CAF1 54928 102 + 1 AAAGCCAAAAGGCAAAGGCAAACGUAAACGCAAUUCAAA--------------A--CCAGGUUGCUAUGCCAUGCUCUUUAGAACUUGUUUCGAGUUUUGAGUCUCUGCUUUUCAAA--U .......(((((((.((((....(((...((((((....--------------.--...))))))..)))........((((((((((...)))))))))))))).)))))))....--. ( -23.00) >consensus AAAGGCAAAAGGCAAGUGCAACUGCGUUUGCCACUGCAG______________GUUGCAGAUUGCUCUGCUAUGCUCUUUACAACUUGCAUCAAGUUGU__UUUGGUGCUUUCUAGCUCU ...(((((..(((((((((....))))))))).((((((.((((.....)))).)))))).)))))..((((.((.((..((((((((...)))))))).....)).))....))))... (-22.04 = -25.44 + 3.40)


| Location | 14,822,742 – 14,822,846 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.44 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -17.38 |
| Energy contribution | -19.38 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14822742 104 - 23771897 AGAACUAGAAAGCACCAAA--ACAACUUGAUACAAGUUGUAAAGAGCAUAGCAGAGCAAUAUGCAAC--------------CUGCAGUGGCAAACGCAGUUGCACUUGCCUUUUGCCUUU ...................--((((((((...))))))))((((.(((....((.((((..((((((--------------.(((.((.....))))))))))).))))))..))))))) ( -27.30) >DroSec_CAF1 57533 104 - 1 AGAGCUAGAAAGCACCAAA--ACAACUUGAUGCAAGUUGUAAAGAGCAUAGCAGAGCAAUCUGCAAC--------------CUGCAGUGGCAAACGCAGUUGCACUUGCCUUUUGCUUUU .......(((((((.....--((((((((...))))))))((((.(((..(((((....)))))...--------------.(((((..((....))..)))))..)))))))))))))) ( -33.90) >DroSim_CAF1 50733 104 - 1 AGAGCUAGAAAGCACCAAA--ACAACUUGAUGCAAGUUGUAAAGAGCAUAGCAGAGCAAUCUGCAAC--------------CUGCAGUGGCAAACGCAGUUGCACUUGCCUUUUGCCUUU ((((.(((((.((......--((((((((...)))))))).(((.(((..(((((....)))))...--------------((((.((.....)))))).))).))))).))))).)))) ( -31.20) >DroYak_CAF1 60844 118 - 1 AGAGCUAAAAAACACCAAA--ACAACUUGAUGGAAGUUGUAAAGAGCAUUGCAGAGCAAUCUGCAACCUGCAACCAGCAACCUGCAGUGGCAAACGCAGUUGCACUUGCCUUUUGCCUUU ...(((.............--(((((((.....))))))).....((((((((((....)))))))..)))....))).....((((.(((((..((....))..)))))..)))).... ( -34.90) >DroAna_CAF1 54928 102 - 1 A--UUUGAAAAGCAGAGACUCAAAACUCGAAACAAGUUCUAAAGAGCAUGGCAUAGCAACCUGG--U--------------UUUGAAUUGCGUUUACGUUUGCCUUUGCCUUUUGGCUUU .--.(..(((.((((((..((((((((.(...((.((((....)))).))((...))...).))--)--------------)))))...(((........))))))))).)))..).... ( -22.60) >consensus AGAGCUAGAAAGCACCAAA__ACAACUUGAUGCAAGUUGUAAAGAGCAUAGCAGAGCAAUCUGCAAC______________CUGCAGUGGCAAACGCAGUUGCACUUGCCUUUUGCCUUU .......(((.(((.......(((((((.....)))))))((((.(((..(((((....)))))..................(((((..((....))..)))))..)))))))))).))) (-17.38 = -19.38 + 2.00)


| Location | 14,822,781 – 14,822,886 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.52 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -19.85 |
| Energy contribution | -20.93 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14822781 105 - 23771897 AAGAUGUGGCCACUUCAGACUUUCAACAUGUGGCUGCCACAGAACUAGAAAGCACCAAA--ACAACUUGAUACAAGUUGUAAAGAGCAUAGCAGAGCAAUAUGCAAC------------- .....(..(((((................)))))..)......................--((((((((...)))))))).....(((((.........)))))...------------- ( -22.99) >DroSec_CAF1 57572 105 - 1 AAGAUGUGGCCACUUCAGACUUUCAACAUGUGGCUGCCACAGAGCUAGAAAGCACCAAA--ACAACUUGAUGCAAGUUGUAAAGAGCAUAGCAGAGCAAUCUGCAAC------------- .....(..(((((................)))))..)......(((....)))......--((((((((...))))))))..........(((((....)))))...------------- ( -29.69) >DroSim_CAF1 50772 105 - 1 AAGAUGUGGCCACUUCAGACUUUCAACAUGUGGCUGCCACAGAGCUAGAAAGCACCAAA--ACAACUUGAUGCAAGUUGUAAAGAGCAUAGCAGAGCAAUCUGCAAC------------- .....(..(((((................)))))..)......(((....)))......--((((((((...))))))))..........(((((....)))))...------------- ( -29.69) >DroYak_CAF1 60884 118 - 1 AAGAUGUGGCCACUUCAGACUUUCAGCAUGUGGCUGCCACAGAGCUAAAAAACACCAAA--ACAACUUGAUGGAAGUUGUAAAGAGCAUUGCAGAGCAAUCUGCAACCUGCAACCAGCAA .....(..(((((................)))))..)......(((.............--(((((((.....))))))).....((((((((((....)))))))..)))....))).. ( -34.09) >DroAna_CAF1 54967 103 - 1 UAGAUGCAGCCACUUCAGACUUCCAACAUGUUGCUGCCACA--UUUGAAAAGCAGAGACUCAAAACUCGAAACAAGUUCUAAAGAGCAUGGCAUAGCAACCUGG--U------------- ..........................((.((((((((((..--...........(((........))).......((((....)))).))))..)))))).)).--.------------- ( -20.10) >consensus AAGAUGUGGCCACUUCAGACUUUCAACAUGUGGCUGCCACAGAGCUAGAAAGCACCAAA__ACAACUUGAUGCAAGUUGUAAAGAGCAUAGCAGAGCAAUCUGCAAC_____________ .....((((((((................))))))))........................(((((((.....)))))))..........(((((....)))))................ (-19.85 = -20.93 + 1.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:41 2006