
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,822,566 – 14,822,682 |
| Length | 116 |
| Max. P | 0.998196 |

| Location | 14,822,566 – 14,822,682 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -28.82 |
| Energy contribution | -30.22 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
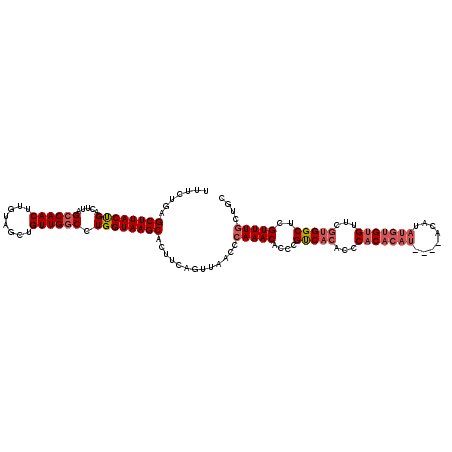
>3L_DroMel_CAF1 14822566 116 + 23771897 UUUCUGAGCUUACUGACUUAGCCAACUUGUAGCUGUUGGCCUGGUAAGCACUUCAGUUAACCCAAACACCCGUCACAACCACACAU----ACAUAUGUGUGUUCGUGGCUCGUUUGCUGC ...((((((((((((.....((((((........)))))).))))))))...))))......(((((....(((((...(((((((----....)))))))...)))))..))))).... ( -35.70) >DroSec_CAF1 57357 116 + 1 UUUCUGAGCUUACUGACUUAGCCAACUUGUAGCUGUUGGCCUGGUAAGCACUUCAGUUAACCCAAACACCCGUCACACCCACACAU----ACAUAUGUGUGUUCGUGGCUCGUUUGAUGC ...((((((((((((.....((((((........)))))).))))))))...))))......(((((....(((((...(((((((----....)))))))...)))))..))))).... ( -36.40) >DroSim_CAF1 50557 116 + 1 UUUCUGAGCUUACUGACUUAGCCAACUUGUAGCUGUUGGCCUGGUAAGCACUUCAGUUAACCCAAACACCCGUCACACCCACACAU----ACAUAUGUGUGUUCGUGGCUCGUUUGCUGC ...((((((((((((.....((((((........)))))).))))))))...))))......(((((....(((((...(((((((----....)))))))...)))))..))))).... ( -35.70) >DroYak_CAF1 60656 120 + 1 UUUCUGAGCUUACCGACUUAGCCAACUUGUAGCUGUUGCCCUGGUAAGCACUUCAGUUAACCCAAACACCCGUCACACCCACACAUAUGUACAUAUGUGUGCUCGUGGCUCGUUUGGUGC ...((((((((((((.....((.(((........)))))..))))))))...)))).....((((((....(((((...(((((((((....)))))))))...)))))..))))))... ( -38.40) >DroAna_CAF1 54762 97 + 1 ------AGCUUACCGUCUUAGCCAACUUGUAGCUGUUGGCCUGGUAAGCACUUUAGUUAUCCCAAACACUCGCC-----CAC------------AUAUGCGUUUGUGGCUUGUUUUUUGG ------.((((((((.....((((((........)))))).))))))))............(((((.((..(((-----.((------------(........))))))..))..))))) ( -30.40) >consensus UUUCUGAGCUUACUGACUUAGCCAACUUGUAGCUGUUGGCCUGGUAAGCACUUCAGUUAACCCAAACACCCGUCACACCCACACAU____ACAUAUGUGUGUUCGUGGCUCGUUUGCUGC .......((((((((.....((((((........)))))).)))))))).............(((((....(((((...(((((((........)))))))...)))))..))))).... (-28.82 = -30.22 + 1.40)

| Location | 14,822,566 – 14,822,682 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.22 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -29.60 |
| Energy contribution | -30.92 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
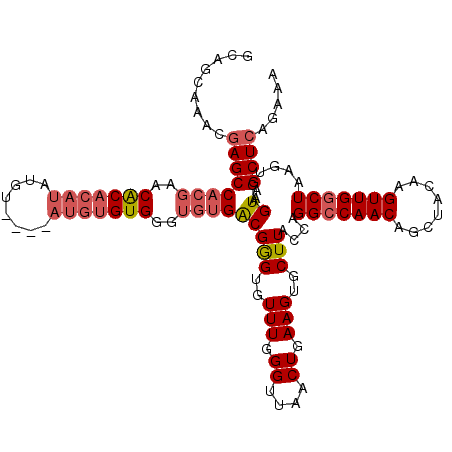
>3L_DroMel_CAF1 14822566 116 - 23771897 GCAGCAAACGAGCCACGAACACACAUAUGU----AUGUGUGGUUGUGACGGGUGUUUGGGUUAACUGAAGUGCUUACCAGGCCAACAGCUACAAGUUGGCUAAGUCAGUAAGCUCAGAAA ....((((((.(.(((((.(((((((....----))))))).))))).)...))))))......((((..((((.((..(((((((........)))))))..)).))))...))))... ( -39.70) >DroSec_CAF1 57357 116 - 1 GCAUCAAACGAGCCACGAACACACAUAUGU----AUGUGUGGGUGUGACGGGUGUUUGGGUUAACUGAAGUGCUUACCAGGCCAACAGCUACAAGUUGGCUAAGUCAGUAAGCUCAGAAA ...(((((((.(.((((..(((((((....----)))))))..)))).)...))))))).....((((..((((.((..(((((((........)))))))..)).))))...))))... ( -41.20) >DroSim_CAF1 50557 116 - 1 GCAGCAAACGAGCCACGAACACACAUAUGU----AUGUGUGGGUGUGACGGGUGUUUGGGUUAACUGAAGUGCUUACCAGGCCAACAGCUACAAGUUGGCUAAGUCAGUAAGCUCAGAAA ....((((((.(.((((..(((((((....----)))))))..)))).)...))))))......((((..((((.((..(((((((........)))))))..)).))))...))))... ( -40.60) >DroYak_CAF1 60656 120 - 1 GCACCAAACGAGCCACGAGCACACAUAUGUACAUAUGUGUGGGUGUGACGGGUGUUUGGGUUAACUGAAGUGCUUACCAGGGCAACAGCUACAAGUUGGCUAAGUCGGUAAGCUCAGAAA ...(((((((.(.((((..(((((((((....)))))))))..)))).)...))))))).....((((...(((((((..(....)(((((.....))))).....)))))))))))... ( -45.80) >DroAna_CAF1 54762 97 - 1 CCAAAAAACAAGCCACAAACGCAUAU------------GUG-----GGCGAGUGUUUGGGAUAACUAAAGUGCUUACCAGGCCAACAGCUACAAGUUGGCUAAGACGGUAAGCU------ ((...(((((.((((((........)------------)).-----)))...))))).))...........(((((((.(((((((........))))))).....))))))).------ ( -31.70) >consensus GCAGCAAACGAGCCACGAACACACAUAUGU____AUGUGUGGGUGUGACGGGUGUUUGGGUUAACUGAAGUGCUUACCAGGCCAACAGCUACAAGUUGGCUAAGUCAGUAAGCUCAGAAA .........((((((((..(((((((........)))))))..))))(((((..(((.((....)).)))..)))....(((((((........)))))))......))..))))..... (-29.60 = -30.92 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:39 2006