
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,821,218 – 14,821,396 |
| Length | 178 |
| Max. P | 0.997205 |

| Location | 14,821,218 – 14,821,316 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -21.38 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14821218 98 + 23771897 GGCGGCGACGCCUUCAGGCCUCACUUUUAUCCUCAUGCCAGGACUCAGUUUCGGAUGCACCGCCUCCACUU-UGGCAUUACUGCCGCAAAACUCUUUGC .(((((((.(((....))).))............(((((((((......)))(((.((...)).)))....-))))))....)))))............ ( -29.00) >DroSec_CAF1 56029 98 + 1 GGCGGCGACGCCUUCAGGCCUCACUUUUAUCCUCAUGCCAGGACUCAGUUUCGGAUGCACCGCCUCCACUU-UGGCAUUACUGCCGCAAAACUCUUUGC .(((((((.(((....))).))............(((((((((......)))(((.((...)).)))....-))))))....)))))............ ( -29.00) >DroSim_CAF1 49218 99 + 1 GGCGGCGACGCCUUCAGGCCUCACUUUUAUCCUCAUGCCAGGACUCAGUUUCGGAUGCACCGCCUCCACUUUUGGCAUUACUGCCACAAAACUCUUUGC (((((.((.(((....))).))............(((((((((......)))(((.((...)).))).....))))))..))))).............. ( -25.90) >DroEre_CAF1 56459 98 + 1 GGCGGCAACGCCUCCAGGCCUCACUUUUAUCCUCAUUGCAGGACUCAGUUUCGGAUACACCGCCUCCACUU-UGCCAUUACUGCCGCAAAACUCUUUGC ((((....))))....(((..........((((......))))........(((.....))).........-.))).........(((((....))))) ( -21.40) >DroYak_CAF1 59302 98 + 1 GGCGGCAACGCCUCCAGGCCUCACUUUUAUCCUCAUUUCAGGACUCAGUUUCGGAUACACCGCCUCCACUU-UGCCAUUACUGCCGCAAAACUCUUUGC ((((....))))....(((..........((((......))))........(((.....))).........-.))).........(((((....))))) ( -21.40) >consensus GGCGGCGACGCCUUCAGGCCUCACUUUUAUCCUCAUGCCAGGACUCAGUUUCGGAUGCACCGCCUCCACUU_UGGCAUUACUGCCGCAAAACUCUUUGC ((((....))))...((((..........((((......)))).........((.....))))))........((((....))))(((((....))))) (-21.38 = -21.98 + 0.60)

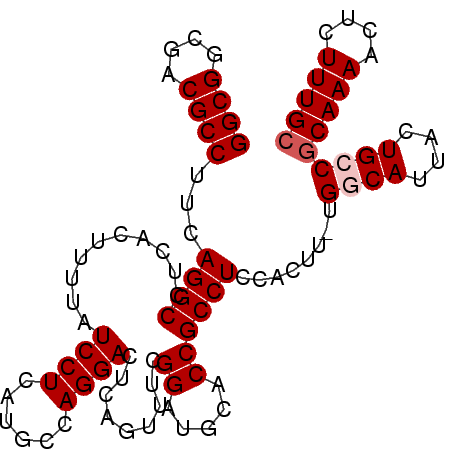

| Location | 14,821,218 – 14,821,316 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.38 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14821218 98 - 23771897 GCAAAGAGUUUUGCGGCAGUAAUGCCA-AAGUGGAGGCGGUGCAUCCGAAACUGAGUCCUGGCAUGAGGAUAAAAGUGAGGCCUGAAGGCGUCGCCGCC (((((....)))))((((....)))).-.......((((((.........(((..(((((......)))))...)))((.(((....))).)))))))) ( -36.60) >DroSec_CAF1 56029 98 - 1 GCAAAGAGUUUUGCGGCAGUAAUGCCA-AAGUGGAGGCGGUGCAUCCGAAACUGAGUCCUGGCAUGAGGAUAAAAGUGAGGCCUGAAGGCGUCGCCGCC (((((....)))))((((....)))).-.......((((((.........(((..(((((......)))))...)))((.(((....))).)))))))) ( -36.60) >DroSim_CAF1 49218 99 - 1 GCAAAGAGUUUUGUGGCAGUAAUGCCAAAAGUGGAGGCGGUGCAUCCGAAACUGAGUCCUGGCAUGAGGAUAAAAGUGAGGCCUGAAGGCGUCGCCGCC .............(((((....)))))........((((((.........(((..(((((......)))))...)))((.(((....))).)))))))) ( -34.00) >DroEre_CAF1 56459 98 - 1 GCAAAGAGUUUUGCGGCAGUAAUGGCA-AAGUGGAGGCGGUGUAUCCGAAACUGAGUCCUGCAAUGAGGAUAAAAGUGAGGCCUGGAGGCGUUGCCGCC (((((....)))))(.((....)).).-.......((((((.........(((..(((((......)))))...)))((.(((....))).)))))))) ( -29.50) >DroYak_CAF1 59302 98 - 1 GCAAAGAGUUUUGCGGCAGUAAUGGCA-AAGUGGAGGCGGUGUAUCCGAAACUGAGUCCUGAAAUGAGGAUAAAAGUGAGGCCUGGAGGCGUUGCCGCC (((((....)))))(.((....)).).-.......((((((.........(((..(((((......)))))...)))((.(((....))).)))))))) ( -29.50) >consensus GCAAAGAGUUUUGCGGCAGUAAUGCCA_AAGUGGAGGCGGUGCAUCCGAAACUGAGUCCUGGCAUGAGGAUAAAAGUGAGGCCUGAAGGCGUCGCCGCC (((((....)))))((((....)))).........((((((.........(((..(((((......)))))...)))((.(((....))).)))))))) (-32.38 = -32.38 + -0.00)

| Location | 14,821,258 – 14,821,356 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 97.56 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -27.22 |
| Energy contribution | -27.38 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14821258 98 - 23771897 AGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGCCA-AAGUGGAGGCGGUGCAUCCGAAACUGAGUCC .(((((...(((((((....((((((((..((((....))))...)))))))).((((....)))).-.)))))))..)))))................ ( -31.00) >DroSec_CAF1 56069 98 - 1 AGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGCCA-AAGUGGAGGCGGUGCAUCCGAAACUGAGUCC .(((((...(((((((....((((((((..((((....))))...)))))))).((((....)))).-.)))))))..)))))................ ( -31.00) >DroSim_CAF1 49258 99 - 1 AGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGUGGCAGUAAUGCCAAAAGUGGAGGCGGUGCAUCCGAAACUGAGUCC .(((((...(((((((...(((((((((..((((....))))...)))))))))((((....))))...)))))))..)))))................ ( -32.50) >DroEre_CAF1 56499 98 - 1 AGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGGCA-AAGUGGAGGCGGUGUAUCCGAAACUGAGUCC ...((((......(((((..((((((((..((((....))))...))))))))..)))))..)))).-....(((..((((.........))))..))) ( -25.40) >DroYak_CAF1 59342 98 - 1 AGCGCUAAACUUCAUUGCUACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGGCA-AAGUGGAGGCGGUGUAUCCGAAACUGAGUCC ...((((......(((((..((((((((..((((....))))...))))))))..)))))..)))).-....(((..((((.........))))..))) ( -26.30) >consensus AGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGCCA_AAGUGGAGGCGGUGCAUCCGAAACUGAGUCC .(((((...(((((((....((((((((..((((....))))...)))))))).((((....))))...)))))))..)))))................ (-27.22 = -27.38 + 0.16)


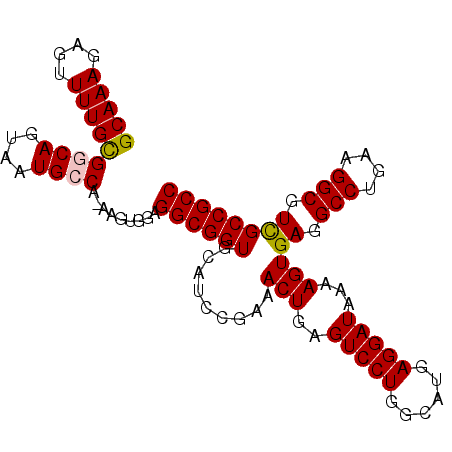
| Location | 14,821,291 – 14,821,396 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -31.25 |
| Consensus MFE | -26.88 |
| Energy contribution | -26.97 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14821291 105 + 23771897 ---------------GCAUUACUGCCGCAAAACUCUUUGCACAUUGGUGCGGAAAUUUUGUUGCAAUGAAGUUUAGCGCUGAGUGCAUGUUGCUCUUGAGCAAAAUUAAUGUAAUUGCUC ---------------(((((((...............(((((.(..((((..((((((..(....)..)))))).))))..))))))..(((((....))))).......)))).))).. ( -29.50) >DroSec_CAF1 56102 105 + 1 ---------------GCAUUACUGCCGCAAAACUCUUUGCACAUUGGUGCGGAAAUUUUGUUGCAAUGAAGUUUAGCGCUGAGUGCAUGUUGCUCUUGAGCAAAAUUAAUGUAAUUGCUC ---------------(((((((...............(((((.(..((((..((((((..(....)..)))))).))))..))))))..(((((....))))).......)))).))).. ( -29.50) >DroSim_CAF1 49292 105 + 1 ---------------GCAUUACUGCCACAAAACUCUUUGCACAUUGGUGCGGAAAUUUUGUUGCAAUGAAGUUUAGCGCUGAGUGCAUGUUGCUCUUGAGCAAAAUUAAUGUAAUUGCUC ---------------(((((((...............(((((.(..((((..((((((..(....)..)))))).))))..))))))..(((((....))))).......)))).))).. ( -29.50) >DroEre_CAF1 56532 105 + 1 ---------------CCAUUACUGCCGCAAAACUCUUUGCACAUUGGUGCGGAAAUUUUGUUGCAAUGAAGUUUAGCGCUGAGUGCAUGUUGCUCUUGAGCAAAAUUAAUGUAAUUGCUC ---------------...........((((.......(((((.(..((((..((((((..(....)..)))))).))))..))))))..(((((....)))))...........)))).. ( -28.40) >DroYak_CAF1 59375 105 + 1 ---------------CCAUUACUGCCGCAAAACUCUUUGCACAUUGGUGCGGAAAUUUUGUAGCAAUGAAGUUUAGCGCUGAGUGCAUGUUGCUCUUGAGCAAAAUUAAUGUAAUUGCUC ---------------...........((((.......(((((.(..((((..((((((..(....)..)))))).))))..))))))..(((((....)))))...........)))).. ( -28.60) >DroAna_CAF1 53562 120 + 1 GCAAGCGGUAGCGCGGCGUUACUGCCGCAAAACUCUUUGCACAUUGGUGCGGAAAUUUUGUUGCAAUGAAGUUUAGCGCCGAGUGCAUGUUGCACUUGAGUAAAAUUAAUGUAAUUGCUC (((((((((((((...))))))))).(((((....)))))((((((((((..((((((..(....)..)))))).)))))((((((.....))))))..........)))))..)))).. ( -42.00) >consensus _______________GCAUUACUGCCGCAAAACUCUUUGCACAUUGGUGCGGAAAUUUUGUUGCAAUGAAGUUUAGCGCUGAGUGCAUGUUGCUCUUGAGCAAAAUUAAUGUAAUUGCUC ...............(((((((...............(((((.(((((((..((((((..(....)..)))))).))))))))))))..(((((....))))).......)))).))).. (-26.88 = -26.97 + 0.09)

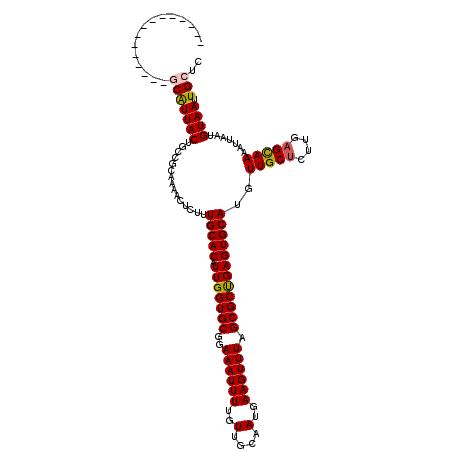

| Location | 14,821,291 – 14,821,396 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.15 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -23.88 |
| Energy contribution | -24.05 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14821291 105 - 23771897 GAGCAAUUACAUUAAUUUUGCUCAAGAGCAACAUGCACUCAGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGC--------------- ((((((...........))))))....(((....((.....))..........(((((..((((((((..((((....))))...))))))))..)))))..)))--------------- ( -26.50) >DroSec_CAF1 56102 105 - 1 GAGCAAUUACAUUAAUUUUGCUCAAGAGCAACAUGCACUCAGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGC--------------- ((((((...........))))))....(((....((.....))..........(((((..((((((((..((((....))))...))))))))..)))))..)))--------------- ( -26.50) >DroSim_CAF1 49292 105 - 1 GAGCAAUUACAUUAAUUUUGCUCAAGAGCAACAUGCACUCAGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGUGGCAGUAAUGC--------------- ((((((...........))))))....(((....((.....))..........(((((.(((((((((..((((....))))...))))))))).)))))..)))--------------- ( -26.90) >DroEre_CAF1 56532 105 - 1 GAGCAAUUACAUUAAUUUUGCUCAAGAGCAACAUGCACUCAGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGG--------------- ((((((...........))))))........(((((.....))..........(((((..((((((((..((((....))))...))))))))..))))).))).--------------- ( -25.70) >DroYak_CAF1 59375 105 - 1 GAGCAAUUACAUUAAUUUUGCUCAAGAGCAACAUGCACUCAGCGCUAAACUUCAUUGCUACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGG--------------- ((((((...........))))))........(((((.....))..........(((((..((((((((..((((....))))...))))))))..))))).))).--------------- ( -26.60) >DroAna_CAF1 53562 120 - 1 GAGCAAUUACAUUAAUUUUACUCAAGUGCAACAUGCACUCGGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAACGCCGCGCUACCGCUUGC ((((....................(((((.....))))).(((((........(((((..((((((((..((((....))))...))))))))..)))))......)))))...)))).. ( -33.74) >consensus GAGCAAUUACAUUAAUUUUGCUCAAGAGCAACAUGCACUCAGCGCUAAACUUCAUUGCAACAAAAUUUCCGCACCAAUGUGCAAAGAGUUUUGCGGCAGUAAUGC_______________ ((((((...........))))))....((.....)).................(((((..((((((((..((((....))))...))))))))..))))).................... (-23.88 = -24.05 + 0.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:35 2006