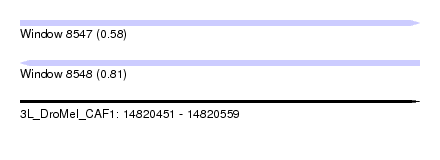
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,820,451 – 14,820,559 |
| Length | 108 |
| Max. P | 0.813222 |

| Location | 14,820,451 – 14,820,559 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -17.64 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
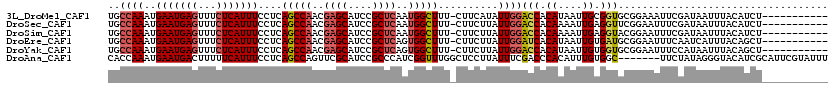
>3L_DroMel_CAF1 14820451 108 + 23771897 -----------AGAUGUAAAUUAUCGAAUUUCCGCACCGCAAUUAUGUGGUCCAAUAUGAAG-AAAGCCAUUGAGCGGAUGCUCGUUGGCUGAGGAAAUGAGAAACUCAUUCAUUUGGCA -----------(((((.......((..(((((((.((((((....)))))).).........-..(((((.(((((....))))).)))))..))))))..))........))))).... ( -31.76) >DroSec_CAF1 55275 108 + 1 -----------AGAUGUAAAUUAUCGAAAUUCCGAACCUCAAUUUUGUGGUCCAAUAAGAAG-AAAGCCAUUGAGCGGAUGCUCGUUGGCUGAGGAAAUGAGAAACUCAUUCAUUUGGCA -----------.((((.....))))......(((((((((..((((((......))))))..-..(((((.(((((....))))).))))))))).((((((...))))))..))))).. ( -30.60) >DroSim_CAF1 48451 108 + 1 -----------AGAUGUAAAUUAUCGAAAUUCCGUACCUCAAUUUUGUGGUCCAAUAAGAAG-AAAGCCAUUGAGCGGAUGCUCGUUGGCUGAGGAAAUGAGAAACUCAUUCAUUUGGCA -----------.((((.....))))......(((..((((..((((((......))))))..-..(((((.(((((....))))).))))))))).((((((...))))))....))).. ( -28.40) >DroEre_CAF1 55703 108 + 1 -----------AGCUGUAAAUGAUUGAAAUUCCGCAUCACAAUUAUGUGAUCCAAUAAGAAG-AAAGCCACUGAGCGGAUGCUCGUUGGCUGAGGAAAUGAGAAACUCAUUCAUUUGGCA -----------.(((..((((((......(((((.((((((....)))))).).........-..(((((.(((((....))))).)))))..))))(((((...)))))))))))))). ( -33.10) >DroYak_CAF1 58517 108 + 1 -----------AGCUGUAAAUUAUGGAAAUUCCGCACCACAAUUAUGUGGUCCAAUAAGAAG-AAAGCCACUGAGCGGAUGCUCGUUGGCUGAGGAAAUGAGAAACUCAUUCAUUUGGCA -----------.((((......(((((..(((((.((((((....)))))).).........-..(((((.(((((....))))).)))))..)))).((((...))))))))).)))). ( -32.20) >DroAna_CAF1 52846 113 + 1 AAAUACGAAUGCGAUGUACCCUAUAGAA-------GCCACAAAUGUGGGUCGAAAUAAGGAGCCAAACCGAUGGGCGGAUGCGAACUGGCUGAGGAAAUGAAAAAGUCAUUCAUUUGGUG .....((((((.((((.(((((.....(-------((((......(((.((........)).)))..(((.....)))........))))).)))...(....).))))))))))))... ( -24.20) >consensus ___________AGAUGUAAAUUAUCGAAAUUCCGCACCACAAUUAUGUGGUCCAAUAAGAAG_AAAGCCAUUGAGCGGAUGCUCGUUGGCUGAGGAAAUGAGAAACUCAUUCAUUUGGCA .........................(((.(((((.(((.((....)).))).)............(((((.(((((....))))).)))))..)))).((((...)))))))........ (-17.64 = -18.53 + 0.89)



| Location | 14,820,451 – 14,820,559 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
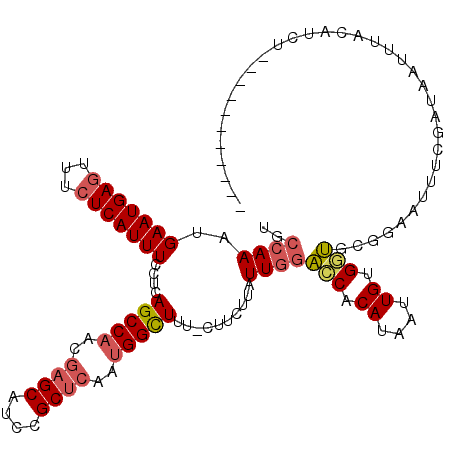
>3L_DroMel_CAF1 14820451 108 - 23771897 UGCCAAAUGAAUGAGUUUCUCAUUUCCUCAGCCAACGAGCAUCCGCUCAAUGGCUUU-CUUCAUAUUGGACCACAUAAUUGCGGUGCGGAAAUUCGAUAAUUUACAUCU----------- ....((((...(((((((((.........(((((..((((....))))..)))))..-.........(.(((.((....)).))).))))))))))...))))......----------- ( -26.90) >DroSec_CAF1 55275 108 - 1 UGCCAAAUGAAUGAGUUUCUCAUUUCCUCAGCCAACGAGCAUCCGCUCAAUGGCUUU-CUUCUUAUUGGACCACAAAAUUGAGGUUCGGAAUUUCGAUAAUUUACAUCU----------- ...........((((((((....((((..(((((..((((....))))..)))))..-.........(((((.((....)).)))))))))....)).)))))).....----------- ( -24.80) >DroSim_CAF1 48451 108 - 1 UGCCAAAUGAAUGAGUUUCUCAUUUCCUCAGCCAACGAGCAUCCGCUCAAUGGCUUU-CUUCUUAUUGGACCACAAAAUUGAGGUACGGAAUUUCGAUAAUUUACAUCU----------- ..((....(((((((...)))))))(((((((((..((((....))))..))))...-.......(((.....)))...)))))...))....................----------- ( -24.50) >DroEre_CAF1 55703 108 - 1 UGCCAAAUGAAUGAGUUUCUCAUUUCCUCAGCCAACGAGCAUCCGCUCAGUGGCUUU-CUUCUUAUUGGAUCACAUAAUUGUGAUGCGGAAUUUCAAUCAUUUACAGCU----------- .((.((((((.((((.((((.........(((((..((((....))))..)))))..-.........(.((((((....)))))).))))).)))).))))))...)).----------- ( -32.60) >DroYak_CAF1 58517 108 - 1 UGCCAAAUGAAUGAGUUUCUCAUUUCCUCAGCCAACGAGCAUCCGCUCAGUGGCUUU-CUUCUUAUUGGACCACAUAAUUGUGGUGCGGAAUUUCCAUAAUUUACAGCU----------- ..((((..(((((((...)))))))....(((((..((((....))))..)))))..-.......))))((((((....))))))((.(((((.....)))))...)).----------- ( -31.90) >DroAna_CAF1 52846 113 - 1 CACCAAAUGAAUGACUUUUUCAUUUCCUCAGCCAGUUCGCAUCCGCCCAUCGGUUUGGCUCCUUAUUUCGACCCACAUUUGUGGC-------UUCUAUAGGGUACAUCGCAUUCGUAUUU ......(((((((................((((((..((.((......))))..))))))((((((...((.((((....)))).-------))..)))))).......))))))).... ( -24.70) >consensus UGCCAAAUGAAUGAGUUUCUCAUUUCCUCAGCCAACGAGCAUCCGCUCAAUGGCUUU_CUUCUUAUUGGACCACAUAAUUGUGGUGCGGAAUUUCGAUAAUUUACAUCU___________ ..((((..(((((((...)))))))....(((((..((((....))))..)))))..........))))(((.((....)).)))................................... (-18.00 = -18.53 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:28 2006