
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,812,271 – 14,812,373 |
| Length | 102 |
| Max. P | 0.961049 |

| Location | 14,812,271 – 14,812,373 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.28 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -12.65 |
| Energy contribution | -12.66 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14812271 102 + 23771897 UCGCCCCAUCCCCGUGCCCCCU----------CCCUACGUAUUGUGCGGAUUUGUGUGCGAGUAUUUUCAAUUAUAUAC--------ACACAUUUGUACACAUAACCACGCAUGCAUAAA ......................----------......((((.(((.((...(((((((((((................--------....)))))))))))...)).)))))))..... ( -20.35) >DroSec_CAF1 47387 103 + 1 UCGCCCCAUCCCCGUGCCUCCA---------CCCCUGCGGAUUGUACGGAUUUGUGUGCGAGUAUUUUCAAUUAUAUAC--------ACACAUUUGUACACAUAACAACGCAUGCAUAAA ..((.......((((((.(((.---------.......)))..))))))...(((((((((((................--------....))))))))))).......))......... ( -23.45) >DroSim_CAF1 40419 103 + 1 UCGCCCCAUCCCGGUGCCUCCA---------CCCCUGCGGAUUUUACGGAUUUGUGUGCGAGUAUUUUCAAUUAUAUAC--------ACACAUUUGUACACAUAACCACGCAUGCAUAAA ..((........((((....))---------))..(((((......)((...(((((((((((................--------....)))))))))))...)).)))).))..... ( -21.25) >DroEre_CAF1 46805 106 + 1 UCGCCCCAUCCCCGUACCCCCAA------CCCCCCCUCGGAUUGUGCGGAUUUGUGUGCGAGUAUUUUCAAUUAUAUAC--------ACACAUUUGUGCACAUGACCACGCAUGCAUAAA (((((.((...((((((..((..------.........))...))))))...)).).))))((((..........))))--------.....((((((((..((....))..)))))))) ( -21.90) >DroYak_CAF1 50415 96 + 1 UCGCCCCCUCUCC----------------CCCUCUUUUGGAUUGUGCGGAUUUGUGUGCGAGUAUUUUCAAUUAUAUAC--------ACACAUUUGUACACAUAACCACGCAUGCAUAAA ..((.........----------------.((......))..((((.((...(((((((((((................--------....)))))))))))...)).)))).))..... ( -20.75) >DroAna_CAF1 45297 110 + 1 UCACCAC----------CACCACCCCUCCUACUCUUAUGGAUUUGUUGUGUGUGUGUGUGAGUAUUUUCAAUUAUACACCCACACACAAACAUCCACACACACAUCCACGGGUGCAUAAA .......----------...(((((............(((((.(((.(((((((((((((.((((........))))...))))))))......)))))))).))))).)))))...... ( -30.72) >consensus UCGCCCCAUCCCCGUGCCCCCA_________CCCCUACGGAUUGUGCGGAUUUGUGUGCGAGUAUUUUCAAUUAUAUAC________ACACAUUUGUACACAUAACCACGCAUGCAUAAA ...........................................(((((....(((((((((((............................)))))))))))......)))))....... (-12.65 = -12.66 + 0.00)

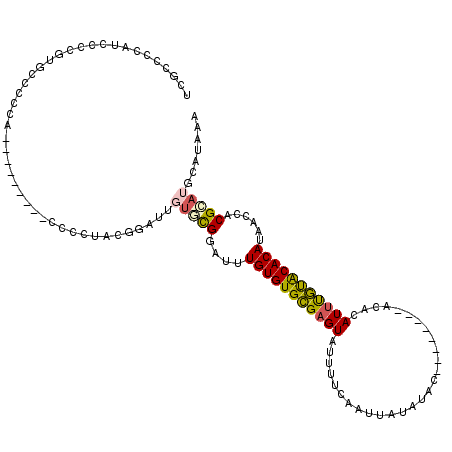
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:19 2006