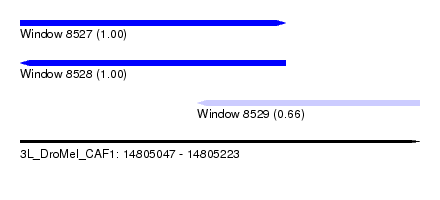
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,805,047 – 14,805,223 |
| Length | 176 |
| Max. P | 0.999410 |

| Location | 14,805,047 – 14,805,164 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.58 |
| SVM RNA-class probability | 0.999410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14805047 117 + 23771897 -UGUAUCUGUGUUUU-CGUGGCAAAGAAGGAAUAAACAAAAUAAAUAAUUCAUAAUGCAAAUAGGCCAAAACACACAGAGACAAGCCG-AAUGCCAAGGCAACGGCGAAAUAAGAGCCCG -(((.(((((((...-..((((.......((((..............))))...((....))..))))....))))))).))).((((-..(((....))).)))).............. ( -26.44) >DroSec_CAF1 40417 118 + 1 -UGUAUCUGUGUUUU-CGUGGCAAAGAAGGAAUAAACAAAAUAAAUAAUUCAUAAUGCAAAUAGGCCAAAACACACAGAGACAAGCCGGAAUGCCAAGGCAACGGCAAAAUAAUAGCCCG -(((.(((((((...-..((((.......((((..............))))...((....))..))))....))))))).))).((((...(((....))).)))).............. ( -27.34) >DroSim_CAF1 33234 118 + 1 -UGUAUCUGUGUUUU-CGUGGCAAAGAAGGAAUAAACAAAAUAAAUAAUUCAUAAUGCAAAUAGGCCAAAACACACAGAGACAAGCCGGAAUGCCAAGGCAACGGCAAAAUAAGAGCCCG -(((.(((((((...-..((((.......((((..............))))...((....))..))))....))))))).))).((((...(((....))).)))).............. ( -27.34) >DroEre_CAF1 39742 118 + 1 GUGUAUCUGUGUUUU-CGUGGCAAAGAAGGAAUAAACAAAAUAAAUAAUUCAUAAUGCAAAUAGGCCAAAACACACAGAGACAAGCCG-AAUGCCAAGGCAACGGCAAAAUAAGAGCCCG .(((.(((((((...-..((((.......((((..............))))...((....))..))))....))))))).))).((((-..(((....))).)))).............. ( -26.64) >DroYak_CAF1 43093 117 + 1 -UGUAUCUGUGUUCU-CGUGGCAAAGAAGGAAUAAACAAAAUAAAUAAUUCAUAAUGCAAAUAGGCCAAAACACACAGAGACAAGCCG-AAUGCCAAGGCAACGGCAAAAUAAGAGCCCG -(((.(((((((...-..((((.......((((..............))))...((....))..))))....))))))).))).((((-..(((....))).)))).............. ( -26.44) >DroAna_CAF1 39264 118 + 1 -UGUAUUUGUGUUUUUGGUGGCAAAGACGGUAUAAACAAAAUAAAUAAUUCAUAAUGCAAAUAGGCCAAAACACACAGAGACAAACCG-AAUGCCAAGGCAACGGAAAAAUAGGAGCUCG -(((.(((((((.(((..((((.......((((.....................))))......))))))).))))))).)))..(((-..(((....))).)))............... ( -22.12) >consensus _UGUAUCUGUGUUUU_CGUGGCAAAGAAGGAAUAAACAAAAUAAAUAAUUCAUAAUGCAAAUAGGCCAAAACACACAGAGACAAGCCG_AAUGCCAAGGCAACGGCAAAAUAAGAGCCCG .(((.(((((((.((...((((.......((((..............))))...((....))..)))).)).))))))).))).((((...(((....))).)))).............. (-24.16 = -24.52 + 0.36)



| Location | 14,805,047 – 14,805,164 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -25.18 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14805047 117 - 23771897 CGGGCUCUUAUUUCGCCGUUGCCUUGGCAUU-CGGCUUGUCUCUGUGUGUUUUGGCCUAUUUGCAUUAUGAAUUAUUUAUUUUGUUUAUUCCUUCUUUGCCACG-AAAACACAGAUACA- ..............((((.(((....)))..-)))).(((.(((((((.(((((((..((..(((..((((.....))))..)))..)).........)))).)-)).))))))).)))- ( -29.20) >DroSec_CAF1 40417 118 - 1 CGGGCUAUUAUUUUGCCGUUGCCUUGGCAUUCCGGCUUGUCUCUGUGUGUUUUGGCCUAUUUGCAUUAUGAAUUAUUUAUUUUGUUUAUUCCUUCUUUGCCACG-AAAACACAGAUACA- ..............((((.(((....)))...)))).(((.(((((((.(((((((..((..(((..((((.....))))..)))..)).........)))).)-)).))))))).)))- ( -28.20) >DroSim_CAF1 33234 118 - 1 CGGGCUCUUAUUUUGCCGUUGCCUUGGCAUUCCGGCUUGUCUCUGUGUGUUUUGGCCUAUUUGCAUUAUGAAUUAUUUAUUUUGUUUAUUCCUUCUUUGCCACG-AAAACACAGAUACA- ..............((((.(((....)))...)))).(((.(((((((.(((((((..((..(((..((((.....))))..)))..)).........)))).)-)).))))))).)))- ( -28.20) >DroEre_CAF1 39742 118 - 1 CGGGCUCUUAUUUUGCCGUUGCCUUGGCAUU-CGGCUUGUCUCUGUGUGUUUUGGCCUAUUUGCAUUAUGAAUUAUUUAUUUUGUUUAUUCCUUCUUUGCCACG-AAAACACAGAUACAC ..............((((.(((....)))..-)))).(((.(((((((.(((((((..((..(((..((((.....))))..)))..)).........)))).)-)).))))))).))). ( -29.40) >DroYak_CAF1 43093 117 - 1 CGGGCUCUUAUUUUGCCGUUGCCUUGGCAUU-CGGCUUGUCUCUGUGUGUUUUGGCCUAUUUGCAUUAUGAAUUAUUUAUUUUGUUUAUUCCUUCUUUGCCACG-AGAACACAGAUACA- ..............((((.(((....)))..-)))).(((.(((((((.((.((((..((..(((..((((.....))))..)))..)).........)))).)-)..))))))).)))- ( -28.90) >DroAna_CAF1 39264 118 - 1 CGAGCUCCUAUUUUUCCGUUGCCUUGGCAUU-CGGUUUGUCUCUGUGUGUUUUGGCCUAUUUGCAUUAUGAAUUAUUUAUUUUGUUUAUACCGUCUUUGCCACCAAAAACACAAAUACA- .((((..........(((.(((....)))..-)))...).)))..(((((((((((......((..(((((((..........)))))))..))....))))....)))))))......- ( -20.32) >consensus CGGGCUCUUAUUUUGCCGUUGCCUUGGCAUU_CGGCUUGUCUCUGUGUGUUUUGGCCUAUUUGCAUUAUGAAUUAUUUAUUUUGUUUAUUCCUUCUUUGCCACG_AAAACACAGAUACA_ ..............((((.(((....)))...)))).(((.(((((((....((((..((..(((..((((.....))))..)))..)).........))))......))))))).))). (-25.18 = -25.52 + 0.33)

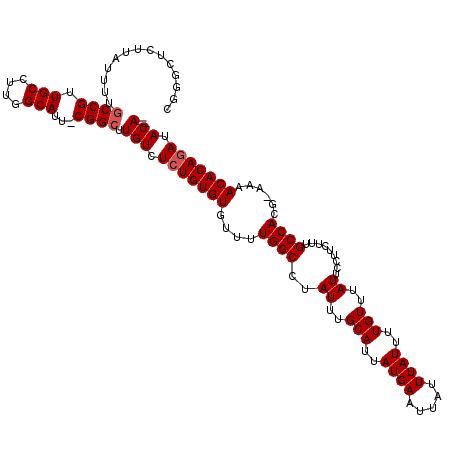
| Location | 14,805,125 – 14,805,223 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.15 |
| Mean single sequence MFE | -41.05 |
| Consensus MFE | -33.03 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14805125 98 - 23771897 --CAUGA-----UGAAGGAGUCCGAG---G---------AGUCAGGAUC--GAGUCCUGGCCGGGUUGCAAUUCCGCCGCCGGGCUCUUAUUUCGCCGUUGCCUUGGCAUU-CGGCUUGU --...((-----...(((((((((.(---(---------.((((((((.--..))))))))((((.......))))))..)))))))))...))((((.(((....)))..-)))).... ( -42.30) >DroSec_CAF1 40495 99 - 1 --CAUGA-----UGGAGGAGUCCGAG---G---------AGUCGGGAUC--GAGUCCUGGCCGGCUUGCAAUUCCGCCGCCGGGCUAUUAUUUUGCCGUUGCCUUGGCAUUCCGGCUUGU --.....-----....((((((((((---(---------...(((.(..--.((.((((((.(((..........))))))))))).......).)))...)))))).)))))....... ( -37.40) >DroSim_CAF1 33312 99 - 1 --CAUGA-----UGGAGGAGUCCGAG---G---------AGUCAGGAUC--GAGUCCUGGCCGGGUUGCAAUUCCGCCGCCGGGCUCUUAUUUUGCCGUUGCCUUGGCAUUCCGGCUUGU --.....-----...(((((((((.(---(---------.((((((((.--..))))))))((((.......))))))..))))))))).....((((.(((....)))...)))).... ( -40.50) >DroEre_CAF1 39821 97 - 1 ---AUGA-----UGAAGGAGUCCGAG---G---------AGUCAGGAUC--GAGUCCUGGCCGGGUUGCAAUUCCGCCGCCGGGCUCUUAUUUUGCCGUUGCCUUGGCAUU-CGGCUUGU ---....-----...(((((((((.(---(---------.((((((((.--..))))))))((((.......))))))..))))))))).....((((.(((....)))..-)))).... ( -41.10) >DroYak_CAF1 43171 107 - 1 --CGUGA-----UGGAGGAGUCCGAG---GAGUCCAAGGAGUCAGGAUC--GAGUCCUGGCCGGGUUGCAAUUUCGCCGCCGGGCUCUUAUUUUGCCGUUGCCUUGGCAUU-CGGCUUGU --.....-----((((....))))((---((((((...(.((((((((.--..))))))))).(((.((......)).))))))))))).....((((.(((....)))..-)))).... ( -42.10) >DroAna_CAF1 39343 109 - 1 GGCAAAAUAGAGUUAAGGACUCUAAGUCCG---------AGCCAGGAUUCGGAGUCCGAGCCGGGUUGCAAUCAUGCCU-CGAGCUCCUAUUUUUCCGUUGCCUUGGCAUU-CGGUUUGU ...(((((((......((((((..(((((.---------.....)))))..))))))((((((((..(((....)))))-)).))))))))))).(((.(((....)))..-)))..... ( -42.90) >consensus __CAUGA_____UGAAGGAGUCCGAG___G_________AGUCAGGAUC__GAGUCCUGGCCGGGUUGCAAUUCCGCCGCCGGGCUCUUAUUUUGCCGUUGCCUUGGCAUU_CGGCUUGU ...............((((((((.................((((((((.....))))))))..(((.((......)).))))))))))).....((((.(((....)))...)))).... (-33.03 = -34.00 + 0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:08 2006