
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,800,225 – 14,800,334 |
| Length | 109 |
| Max. P | 0.708384 |

| Location | 14,800,225 – 14,800,334 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.73 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -21.68 |
| Energy contribution | -21.05 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14800225 109 + 23771897 UCAUUGGGUUCCUUUGAGCCAGCUAAACAAUGUUCCCUGUCAGCAGAAC---UCGUACUACCAUAUA--------CUAGUCCCAACGACAGGCAGCACUUUUUGUUGGCCACCGCUCGAA ...((((((.....((.((((((.(((...((((.((((((....(.((---(.(((........))--------).))).)....)))))).))))..))).))))))))..)))))). ( -28.80) >DroSec_CAF1 35672 109 + 1 UCAUUGGGCUCCUUUGAGCCAGCUAAACAAUGUUCCCUGUCAGCAGAAC---UCGUACUACCAUAUA--------CUAGUCCCAACGACAGGCGGCACUUUUUGUUGGCCACCGCUCGAA ...((((((.....((.((((((.(((...(((..((((((....(.((---(.(((........))--------).))).)....))))))..)))..))).))))))))..)))))). ( -30.80) >DroSim_CAF1 28633 105 + 1 UCAUUGGGCUCCUUUGAGCCAGCUAAACAAUGUUCCCUGUCAGCAGAAC---UAGU----CAGCAUA--------CUAGUCCCAACGACAGGCAGCACUUUUUGUUGGCCACCGCUCGAA ...((((((.....((.((((((.(((...((((.((((((....(.((---((((----......)--------))))).)....)))))).))))..))).))))))))..)))))). ( -33.70) >DroEre_CAF1 35286 115 + 1 CCAUUG-GUUCCUUUGAGCCAGCUAAACAAUGUUCCCUGUCAGCAGAAC---CCGCACUACCAUAUACCA-UAUAGUAGUCCCAACGACAGGCAGCACUUUUUGUUAGCCGCCGCUCGAA ......-......((((((..(((((.(((((((.((((((.((.....---..))(((((.((((...)-))).)))))......)))))).))))....))))))))....)))))). ( -31.00) >DroYak_CAF1 37567 117 + 1 UCAUUGGGCUCCUUUGAGCCAGCUAAACAAUGUUCCCUGUCAGCAGAAC---UCGUACUACCAUAUAGUAGUAUAGUGGUUCCACCGACAGGCAGCACUUUUUGUUAGCCGCCGCUCGAA ...((((((((....))))).(((((.(((((((.((((((.(..((((---(.(((((((......)))))))...)))))..).)))))).))))....)))))))).......))). ( -38.10) >DroAna_CAF1 34039 120 + 1 UCAUUUGGUGCCUUUGAGCCAGCUAAACAAUGUUCCUUGUCAGCAGGACUACUCGUUCUAGUAGUUGUUCUUGCACUCCCUCCAAUGGCAGGCAGCAGUUUUUGCUAGCCACCGCUCAAA .............((((((..((((.((((......))))..(((((((((((......)))))....))))))...........)))).((((((((...))))).)))...)))))). ( -31.90) >consensus UCAUUGGGCUCCUUUGAGCCAGCUAAACAAUGUUCCCUGUCAGCAGAAC___UCGUACUACCAUAUA________CUAGUCCCAACGACAGGCAGCACUUUUUGUUAGCCACCGCUCGAA .............((((((..(((((.(((((((.((((((.............................................)))))).))))....))))))))....)))))). (-21.68 = -21.05 + -0.64)

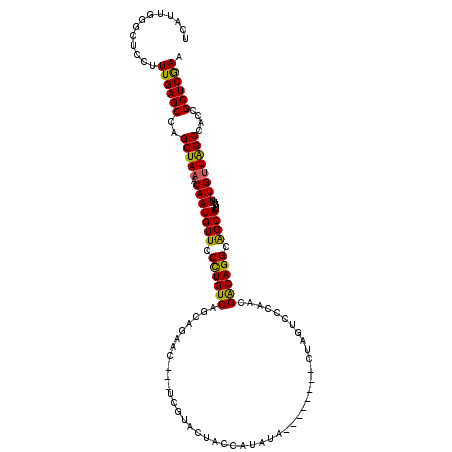
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:04 2006