
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,799,407 – 14,799,635 |
| Length | 228 |
| Max. P | 0.866993 |

| Location | 14,799,407 – 14,799,526 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.05 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.64 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14799407 119 + 23771897 UCAUGUAAAAAAUAAAUUUACAUUUGACCAUAAAAUGAUUUCAAACAGCAUUUAAGGCUGAAUUUACAAUAAUUCCCUUCUGCUAUACGCACUCGAUAAAUGCUUGAUUAAAUCAUGUU ..(((((((.......)))))))...........(((((((.....(((((((((((..(((((......))))))))..(((.....))).....)))))))).....)))))))... ( -20.30) >DroSec_CAF1 34864 119 + 1 UCACGUAAAAAAUAAAUUUACAUUUGCCCAUAAAAUGAUGUCAAACAGCAUUUAAGGCUGAAUUUACAAUAAUUCCCGUCUGCUAUACGCACUCGAUAAAUGCUAGUUUAAAUCAUGUU ....(((((.......))))).............(((((...((((((((((((((((.(((((......)))))..))))((.....))......)))))))).))))..)))))... ( -22.40) >DroSim_CAF1 27807 119 + 1 UCACGUAAAAAAUAAAUUUAUAUUUGCCCAUAAAAUGAUUUCAAACAGCAUUUAUGUCUGAAUUUACAAUAAUACCCGUCUGCUAUACGCACUCGAUAAAUGCUAGAUUAAAUCAUGUC ..................................(((((((.....(((((((((((........)).........((..(((.....)))..))))))))))).....)))))))... ( -16.60) >DroEre_CAF1 34462 117 + 1 UCACGUAAUAAAUAAAUUUUCAUUUACACAUUAUAUGAUUUCAAACAGCAUUUACCGCUGAAUUUACAAUAAUUCCCUUCUGCUAUACGCACUCGGCAAAUGCUUGAUUA--UCAUGUU ....((((...............)))).....(((((((.(((...(((((((.(((..(((((......))))).....(((.....)))..))).))))))))))..)--)))))). ( -20.96) >DroYak_CAF1 36765 106 + 1 UCACGUAUUAAAUGAGUU-------------UAUAUGAUUUCAAACAGCAUUUAACGUUGAAUUUACAAUAAUUCCCUUCUGCUAUACGCACUCGAUAAAUGCUUGAUUAAAUCAUGUU ..................-------------.(((((((((.....((((((((.((..(((((......))))).....(((.....)))..)).)))))))).....))))))))). ( -18.90) >consensus UCACGUAAAAAAUAAAUUUACAUUUGCCCAUAAAAUGAUUUCAAACAGCAUUUAAGGCUGAAUUUACAAUAAUUCCCUUCUGCUAUACGCACUCGAUAAAUGCUUGAUUAAAUCAUGUU ..................................(((((((.....((((((((.....(((((......))))).....(((.....))).....)))))))).....)))))))... (-12.64 = -13.64 + 1.00)

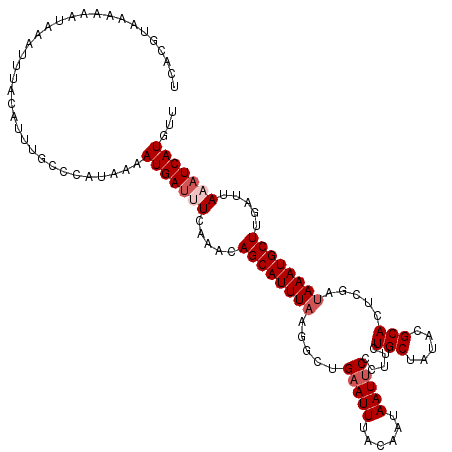

| Location | 14,799,407 – 14,799,526 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 86.05 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -16.66 |
| Energy contribution | -17.14 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14799407 119 - 23771897 AACAUGAUUUAAUCAAGCAUUUAUCGAGUGCGUAUAGCAGAAGGGAAUUAUUGUAAAUUCAGCCUUAAAUGCUGUUUGAAAUCAUUUUAUGGUCAAAUGUAAAUUUAUUUUUUACAUGA ...((((((...((((((((((...))))))..((((((.(((((((((......)))))..))))...))))))))))))))))...........(((((((.......))))))).. ( -25.10) >DroSec_CAF1 34864 119 - 1 AACAUGAUUUAAACUAGCAUUUAUCGAGUGCGUAUAGCAGACGGGAAUUAUUGUAAAUUCAGCCUUAAAUGCUGUUUGACAUCAUUUUAUGGGCAAAUGUAAAUUUAUUUUUUACGUGA ...(((((((((((.(((((((((((..(((.....)))..)))(((((......))))).....)))))))))))))).)))))...........(((((((.......))))))).. ( -26.90) >DroSim_CAF1 27807 119 - 1 GACAUGAUUUAAUCUAGCAUUUAUCGAGUGCGUAUAGCAGACGGGUAUUAUUGUAAAUUCAGACAUAAAUGCUGUUUGAAAUCAUUUUAUGGGCAAAUAUAAAUUUAUUUUUUACGUGA ...(((((((....((((((((((((..(((.....)))..))).......(((........))))))))))))....))))))).(((((...(((((......)))))....))))) ( -21.60) >DroEre_CAF1 34462 117 - 1 AACAUGA--UAAUCAAGCAUUUGCCGAGUGCGUAUAGCAGAAGGGAAUUAUUGUAAAUUCAGCGGUAAAUGCUGUUUGAAAUCAUAUAAUGUGUAAAUGAAAAUUUAUUUAUUACGUGA .......--...((((((((((((((..(((.....))).....(((((......)))))..)))))))))))...)))..(((..(((((.((((((....)))))).)))))..))) ( -31.70) >DroYak_CAF1 36765 106 - 1 AACAUGAUUUAAUCAAGCAUUUAUCGAGUGCGUAUAGCAGAAGGGAAUUAUUGUAAAUUCAACGUUAAAUGCUGUUUGAAAUCAUAUA-------------AACUCAUUUAAUACGUGA ...(((((((.....((((((((.((..(((.....))).....(((((......)))))..)).)))))))).....)))))))...-------------.................. ( -22.20) >consensus AACAUGAUUUAAUCAAGCAUUUAUCGAGUGCGUAUAGCAGAAGGGAAUUAUUGUAAAUUCAGCCUUAAAUGCUGUUUGAAAUCAUUUUAUGGGCAAAUGUAAAUUUAUUUUUUACGUGA ...(((((((.....((((((((((...(((.....)))...))(((((......))))).....)))))))).....))))))).................................. (-16.66 = -17.14 + 0.48)

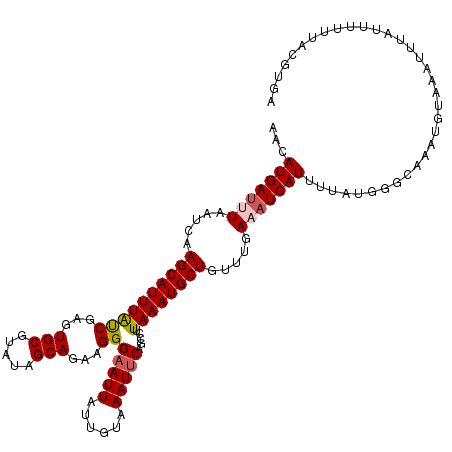

| Location | 14,799,526 – 14,799,635 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -25.21 |
| Consensus MFE | -16.14 |
| Energy contribution | -17.58 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14799526 109 + 23771897 U--AUAUCA---------CUCGACGAAAAAAUAAUCGACGACUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAGCCAC .--......---------.(((.(((........))).))).((((((((.((....(((((((((((.....))))))))))).....)).))))))))(((........)))...... ( -28.90) >DroSec_CAF1 34983 109 + 1 C--AUAUGA---------CUCGACGAAAAAAUAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAGCCAC .--......---------.(((.(((........))).))).((((((((.((....(((((((((((.....))))))))))).....)).))))))))(((........)))...... ( -29.10) >DroSim_CAF1 27926 109 + 1 U--AUAUCA---------CUCGACGAAAAAAUAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAUUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAGCCAC .--......---------.(((.(((........))).))).((((((((.((....(((((((((((.....))))))))))).....)).))))))))(((........)))...... ( -29.10) >DroEre_CAF1 34579 120 + 1 UUUUUAUCAUAGACCAAAAAAAACACAAAAAAAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAAAGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAGCCAC ..........................................((((((((.((....(((((((..((.....))..))))))).....)).))))))))(((........)))...... ( -20.50) >DroYak_CAF1 36871 109 + 1 U--AUAUCA---------AUCGACAGAGAAAAAAACGACGAUUUAAAGUCCUCGAAUGGAUUACAGUGUUGAACAAAGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAGCCAC .--......---------........................((((((((.((....(((((((..((.....))..))))))).....)).))))))))(((........)))...... ( -20.50) >DroAna_CAF1 33257 106 + 1 U--CCAUCA---------GUCAGCAUAUAAAC--UGGCAGCUUUAA-CCGGAACAUUGGCUUAUAGGUUUUUACAAUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUAUAGCGGACCAC .--.(((((---------(((((........)--))))..((((((-..(((((((((...............))))))..))))))))).......)))))(((......)))...... ( -23.16) >consensus U__AUAUCA_________CUCGACAAAAAAAUAAUCGACGAUUUAAAAUCCUCGAAUGGAUUACAGUGUUGAACAAUGUAAUCCUUAAAGAAGAUUUUGAUGCGCAUUACAGCAAGCCAC ..........................................((((((((.((....((((((((.((.....)).)))))))).....)).))))))))(((........)))...... (-16.14 = -17.58 + 1.45)

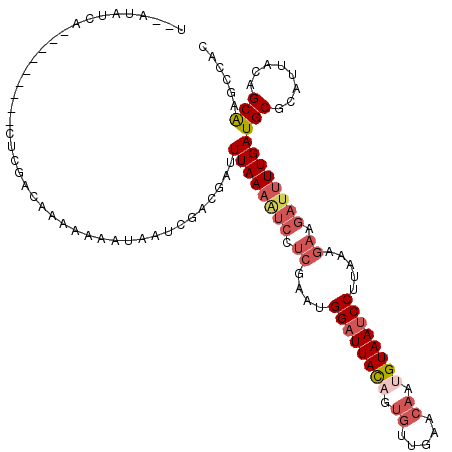
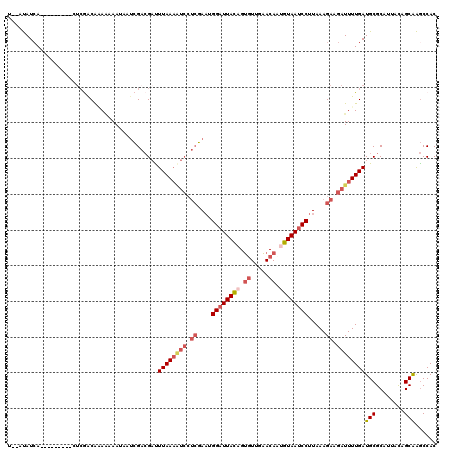
| Location | 14,799,526 – 14,799,635 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.66 |
| Mean single sequence MFE | -25.99 |
| Consensus MFE | -16.09 |
| Energy contribution | -17.48 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14799526 109 - 23771897 GUGGCUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAGUCGUCGAUUAUUUUUUCGUCGAG---------UGAUAU--A ...((..((......))))((((((((((((.....((((((((.((.....)).))))))))....))))))))).(.(((.(((........))).))).---------))))..--. ( -29.00) >DroSec_CAF1 34983 109 - 1 GUGGCUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGAG---------UCAUAU--G (((((..((......)).....(((((((((.....((((((((.((.....)).))))))))....)))))))))...(((.(((........))).))))---------))))..--. ( -31.10) >DroSim_CAF1 27926 109 - 1 GUGGCUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGAG---------UGAUAU--A ...((..((......))))((((((((((((.....((((((((.((.....)).))))))))....)))))))))...(((.(((........))).))).---------.)))..--. ( -28.70) >DroEre_CAF1 34579 120 - 1 GUGGCUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACUUUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUUUUUUUGUGUUUUUUUUGGUCUAUGAUAAAAA ..((((.........(((((..((((((........(((((((..((.....))..)))))))....((.(((.....))).))))))))...))))).......))))........... ( -26.09) >DroYak_CAF1 36871 109 - 1 GUGGCUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACUUUGUUCAACACUGUAAUCCAUUCGAGGACUUUAAAUCGUCGUUUUUUUCUCUGUCGAU---------UGAUAU--A ...((..((......))))((((((.(((((.....(((((((..((.....))..)))))))....))))).))).(((((.((..........)).))))---------))))..--. ( -20.50) >DroAna_CAF1 33257 106 - 1 GUGGUCCGCUAUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAUUGUAAAAACCUAUAAGCCAAUGUUCCGG-UUAAAGCUGCCA--GUUUAUAUGCUGAC---------UGAUGG--A .((((.(((......))).)))).......(((((.(((..((((((...............)))))))))..-.)))))((..((--(((........)))---------))..))--. ( -20.56) >consensus GUGGCUUGCUGUAAUGCGCAUCAAAAUCUUCUUUAAGGAUUACAAUGUUCAACACUGUAAUCCAUUCGAGGAUUUUAAAUCGUCGAUUAUUUUUUCGUCGAG_________UGAUAU__A ..(((.(((......)))....(((((((((.....((((((((.((.....)).))))))))....)))))))))....................)))..................... (-16.09 = -17.48 + 1.39)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:49:03 2006