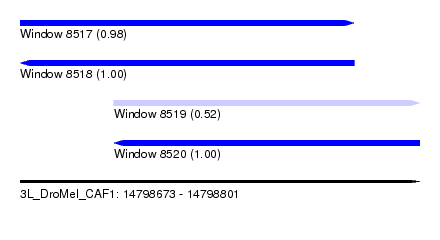
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,798,673 – 14,798,801 |
| Length | 128 |
| Max. P | 0.996253 |

| Location | 14,798,673 – 14,798,780 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -29.66 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.66 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14798673 107 + 23771897 ----------CCUUAGGCCUCGUACAGCUAUUUCAAUGCUACUCGAACUGCCACCCAAAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCGAGUAUUUGGCUUU--- ----------....(((((..(((((((.........)))..(((((.(((((.......(((((((........)))))))....(.....).))))).)))))))))..))))).--- ( -28.40) >DroSec_CAF1 34141 107 + 1 ----------CCUUAGGCCUCGUACAGCUAUAUCAAUGCUACUCGAACUGCCACCCAAAAUUGGUGGAUUCCCUCCCAUUAAAUUAGAUGACGAUGGCAUUUCGAGUAUUUGGCUUU--- ----------....(((((..(((((((.........)))..(((((.(((((.........((.((....)).))((((......))))....))))).)))))))))..))))).--- ( -29.80) >DroEre_CAF1 33610 107 + 1 ----------CCUUAGGCCUUGUAGAGCCAUUUCAAUGCUACUCGAACCGCCACCCGAAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCGAGUAUUUGGCUUU--- ----------.............(((((((.((((((((((.(((..((((((........)))))).........(((........))).))))))))))..)))....)))))))--- ( -26.50) >DroYak_CAF1 35961 107 + 1 ----------CCUUAGGCCUUGUAGAGCAAUUUCAAUGCUCCUCGAACUGCCACCCAAAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCGAGUAUUUGGCUUU--- ----------....(((((..((((((((.......)))))((((((.(((((.......(((((((........)))))))....(.....).))))).)))))))))..))))).--- ( -32.10) >DroAna_CAF1 32465 120 + 1 AGCCAGGGGUCUUUAGGCCUCGUAGAGCUAUUUCAAUGCUACUCGAACUGCCACCCCGAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCAAGCAUUUGGUUUUCUG (((((((((((....)))))).....(((.....(((((((.(((.....(((((.......))))).........(((........))).))))))))))...)))...)))))..... ( -31.50) >consensus __________CCUUAGGCCUCGUAGAGCUAUUUCAAUGCUACUCGAACUGCCACCCAAAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCGAGUAUUUGGCUUU___ ..............(((((..(((.(((.........))).((((((.(((((.........((.((....)).))(((........)))....))))).)))))))))..))))).... (-24.22 = -24.66 + 0.44)



| Location | 14,798,673 – 14,798,780 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.86 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14798673 107 - 23771897 ---AAAGCCAAAUACUCGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUUUGGGUGGCAGUUCGAGUAGCAUUGAAAUAGCUGUACGAGGCCUAAGG---------- ---...(((...((((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))))((.........)).......)))......---------- ( -32.30) >DroSec_CAF1 34141 107 - 1 ---AAAGCCAAAUACUCGAAAUGCCAUCGUCAUCUAAUUUAAUGGGAGGGAAUCCACCAAUUUUGGGUGGCAGUUCGAGUAGCAUUGAUAUAGCUGUACGAGGCCUAAGG---------- ---...(((...((((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))))((.........)).......)))......---------- ( -31.10) >DroEre_CAF1 33610 107 - 1 ---AAAGCCAAAUACUCGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUUCGGGUGGCGGUUCGAGUAGCAUUGAAAUGGCUCUACAAGGCCUAAGG---------- ---.....(((.((((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))))...)))....((((......)))).....---------- ( -32.30) >DroYak_CAF1 35961 107 - 1 ---AAAGCCAAAUACUCGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUUUGGGUGGCAGUUCGAGGAGCAUUGAAAUUGCUCUACAAGGCCUAAGG---------- ---...(((.....((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))(((((.......))))).....)))......---------- ( -36.50) >DroAna_CAF1 32465 120 - 1 CAGAAAACCAAAUGCUUGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUCGGGGUGGCAGUUCGAGUAGCAUUGAAAUAGCUCUACGAGGCCUAAAGACCCCUGGCU ........(((.((((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))))...)))..............((((...........)))) ( -30.10) >consensus ___AAAGCCAAAUACUCGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUUUGGGUGGCAGUUCGAGUAGCAUUGAAAUAGCUCUACGAGGCCUAAGG__________ ......(((...((((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))))((.........)).......)))................ (-29.90 = -29.86 + -0.04)



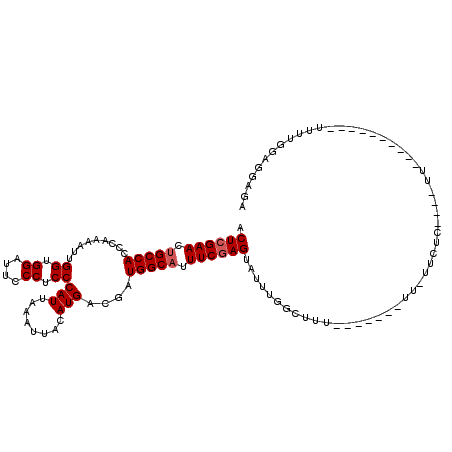
| Location | 14,798,703 – 14,798,801 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -17.12 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14798703 98 + 23771897 ACUCGAACUGCCACCCAAAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCGAGUAUUUGGCUUU-------UU-UUUUC----UU----------UUUUGGAGGAGA (((((((.(((((.......(((((((........)))))))....(.....).))))).)))))))..........-------((-(((((----(.----------....)))))))) ( -24.40) >DroSec_CAF1 34171 96 + 1 ACUCGAACUGCCACCCAAAAUUGGUGGAUUCCCUCCCAUUAAAUUAGAUGACGAUGGCAUUUCGAGUAUUUGGCUUU-------U---UUUC----CU----------UUUUGGAGGAGA (((((((.(((((.........((.((....)).))((((......))))....))))).)))))))........((-------(---((((----(.----------....)))))))) ( -28.20) >DroEre_CAF1 33640 103 + 1 ACUCGAACCGCCACCCGAAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCGAGUAUUUGGCUUU-------UUUUUCUCUUUUUU----------UUUUGUGGGAGA ....(((((((((........)))))).))).(((((((.(((......(((((.......)))(((.....)))..-------......))......----------))).))))))). ( -23.20) >DroYak_CAF1 35991 97 + 1 CCUCGAACUGCCACCCAAAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCGAGUAUUUGGCUUU-------UU-UCCUCU---------------UUUUGUCGGAGA .((((((.(((((.......(((((((........)))))))....(.....).))))).))))))..((((((...-------..-......---------------....)))))).. ( -22.06) >DroAna_CAF1 32505 120 + 1 ACUCGAACUGCCACCCCGAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCAAGCAUUUGGUUUUCUGUUUUUUUUCCUCCUUUUUUUUUUUGUAUUUUGGGAGGAGC ....(((.(((((.......(((((((........)))))))....(.....).))))).))).......................((((((((.................)))))))). ( -23.53) >consensus ACUCGAACUGCCACCCAAAAUUGGUGGAUUCCCUCCCAUUAAAUUACAUGACGAUGGCAUUUCGAGUAUUUGGCUUU_______UU_UUCUC____UU__________UUUUGGAGGAGA .((((((.(((((.........((.((....)).))(((........)))....))))).))))))...................................................... (-17.12 = -17.52 + 0.40)

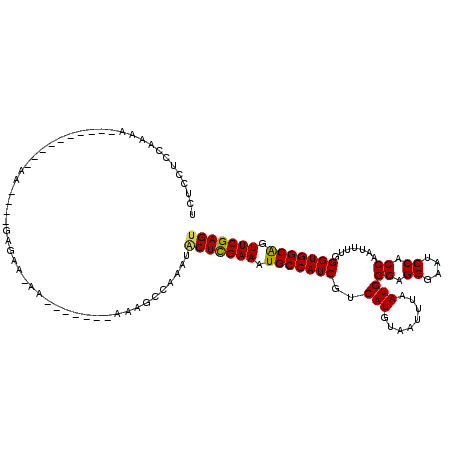
| Location | 14,798,703 – 14,798,801 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -27.21 |
| Consensus MFE | -24.22 |
| Energy contribution | -23.98 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14798703 98 - 23771897 UCUCCUCCAAAA----------AA----GAAAA-AA-------AAAGCCAAAUACUCGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUUUGGGUGGCAGUUCGAGU ............----------..----.....-..-------..........(((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))) ( -25.20) >DroSec_CAF1 34171 96 - 1 UCUCCUCCAAAA----------AG----GAAA---A-------AAAGCCAAAUACUCGAAAUGCCAUCGUCAUCUAAUUUAAUGGGAGGGAAUCCACCAAUUUUGGGUGGCAGUUCGAGU ..((((......----------))----))..---.-------..........(((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))) ( -27.00) >DroEre_CAF1 33640 103 - 1 UCUCCCACAAAA----------AAAAAAGAGAAAAA-------AAAGCCAAAUACUCGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUUCGGGUGGCGGUUCGAGU ((((........----------......))))....-------..........(((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))) ( -25.84) >DroYak_CAF1 35991 97 - 1 UCUCCGACAAAA---------------AGAGGA-AA-------AAAGCCAAAUACUCGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUUUGGGUGGCAGUUCGAGG ..(((.......---------------...)))-..-------...........((((((.(((((((..(((........)))((.((....)).)).......))))))).)))))). ( -26.90) >DroAna_CAF1 32505 120 - 1 GCUCCUCCCAAAAUACAAAAAAAAAAAGGAGGAAAAAAAACAGAAAACCAAAUGCUUGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUCGGGGUGGCAGUUCGAGU ..((((((...................))))))....................(((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))) ( -31.11) >consensus UCUCCUCCAAAA__________AA____GAGAA_AA_______AAAGCCAAAUACUCGAAAUGCCAUCGUCAUGUAAUUUAAUGGGAGGGAAUCCACCAAUUUUGGGUGGCAGUUCGAGU .....................................................(((((((.(((((((..(((........)))((.((....)).)).......))))))).))))))) (-24.22 = -23.98 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:59 2006