
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,789,715 – 14,789,933 |
| Length | 218 |
| Max. P | 0.999962 |

| Location | 14,789,715 – 14,789,826 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.57 |
| Mean single sequence MFE | -37.39 |
| Consensus MFE | -31.51 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14789715 111 + 23771897 CAAUAUCCCCAUACACGCAACAC----ACACUCAAACAC--ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC--- ...............((((((((----(.......((.(--.(((((((......(((((((((((.....)))))))))))))))))).).)))))))))))..............--- ( -37.41) >DroSec_CAF1 32881 111 + 1 CAAUAUCCCCAUAUACGCAACAC----ACACUCAAACAC--ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC--- ...............((((((((----(.......((.(--.(((((((......(((((((((((.....)))))))))))))))))).).)))))))))))..............--- ( -37.41) >DroSim_CAF1 25132 111 + 1 CAAUAUCCCCAUACACGCAACAC----ACACUCAAACAC--ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC--- ...............((((((((----(.......((.(--.(((((((......(((((((((((.....)))))))))))))))))).).)))))))))))..............--- ( -37.41) >DroEre_CAF1 32288 115 + 1 CAAUAUCCCCAUACAGGCAACACACACACACUCAAACAC--ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC--- ...............((.((((((.(((((.....((.(--.(((((((......(((((((((((.....)))))))))))))))))).).))))))))))).)).))........--- ( -36.40) >DroYak_CAF1 34659 111 + 1 CAAUAUCCCCAUACACGCAACAC----ACACUCAAACAC--ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC--- ...............((((((((----(.......((.(--.(((((((......(((((((((((.....)))))))))))))))))).).)))))))))))..............--- ( -37.41) >DroAna_CAF1 31349 115 + 1 CAAUAUCCCCACGCACACACC-G----ACACUCAUACACAUACGGGUGACUGUGGGACAUGACAGCUUUCGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUCA ............((....((.-(----(...(((((((((.(((.(((((((...(((((((((((.....))))))))))))))).))).))))))).)))))..)).))....))... ( -38.30) >consensus CAAUAUCCCCAUACACGCAACAC____ACACUCAAACAC__ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC___ .........((((.(((((((...............(......)....((.(((((((((((((((.....)))))))))))....)))))))))))))))))................. (-31.51 = -31.60 + 0.09)

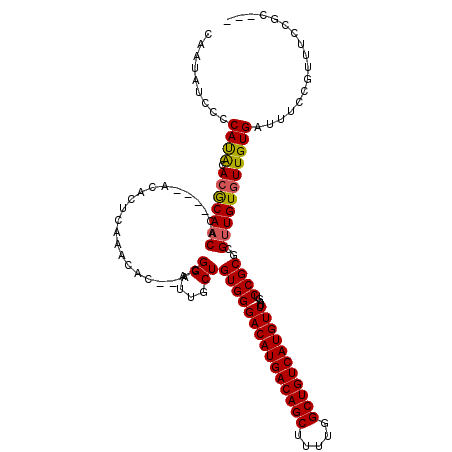
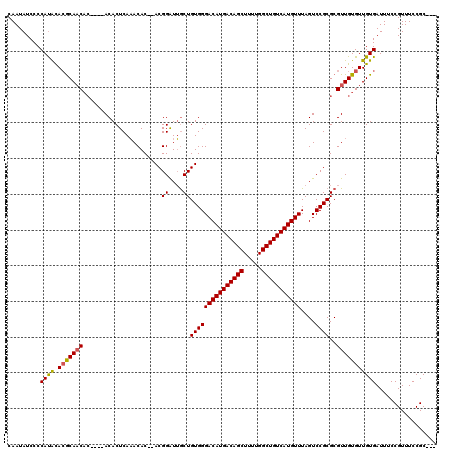
| Location | 14,789,750 – 14,789,865 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.43 |
| Mean single sequence MFE | -43.53 |
| Consensus MFE | -40.06 |
| Energy contribution | -39.78 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14789750 115 + 23771897 -ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC----UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGC -..(((((((((((((((((((((((.....))))))))))).......((((......))))..........))))----.))))))))(((((.((....)).))))).......... ( -43.40) >DroSec_CAF1 32916 115 + 1 -ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC----UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGC -..(((((((((((((((((((((((.....))))))))))).......((((......))))..........))))----.))))))))(((((.((....)).))))).......... ( -43.40) >DroSim_CAF1 25167 115 + 1 -ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC----UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGC -..(((((((((((((((((((((((.....))))))))))).......((((......))))..........))))----.))))))))(((((.((....)).))))).......... ( -43.40) >DroEre_CAF1 32327 115 + 1 -ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC----UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGC -..(((((((((((((((((((((((.....))))))))))).......((((......))))..........))))----.))))))))(((((.((....)).))))).......... ( -43.40) >DroYak_CAF1 34694 115 + 1 -ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC----UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGC -..(((((((((((((((((((((((.....))))))))))).......((((......))))..........))))----.))))))))(((((.((....)).))))).......... ( -43.40) >DroAna_CAF1 31384 120 + 1 UACGGGUGACUGUGGGACAUGACAGCUUUCGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGCUCACUGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGC (((((....)))))((((((((((((.....))))))))))))...(((((((((.(((((.((((.((((........)).)).))))))))))))))..))))............... ( -44.20) >consensus _ACGGAUUGCUGUGGGACAUGACAGCUUUUGGCUGUCAUGUUUAGUCCGCGCGUUGUGUUGUGAUUUCCGUUUCCGC____UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGC ...(((..(((((.((((((((((((.....)))))))))))).......(((((.(((((.((((.(((...........))).))))))))))))))..)))))..)))......... (-40.06 = -39.78 + -0.28)

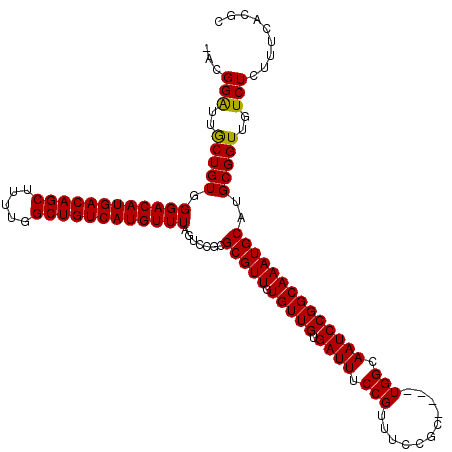
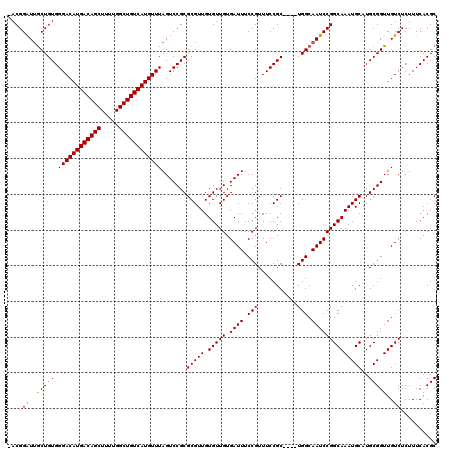
| Location | 14,789,750 – 14,789,865 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.43 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -36.37 |
| Energy contribution | -36.57 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.92 |
| SVM RNA-class probability | 0.999962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14789750 115 - 23771897 GCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA----GCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGU- (((((...(.......).)))))......((((((((..----.((....))..................(.((.....((((((((((.....)))))))))).)).).)))))))).- ( -37.80) >DroSec_CAF1 32916 115 - 1 GCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA----GCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGU- (((((...(.......).)))))......((((((((..----.((....))..................(.((.....((((((((((.....)))))))))).)).).)))))))).- ( -37.80) >DroSim_CAF1 25167 115 - 1 GCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA----GCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGU- (((((...(.......).)))))......((((((((..----.((....))..................(.((.....((((((((((.....)))))))))).)).).)))))))).- ( -37.80) >DroEre_CAF1 32327 115 - 1 GCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA----GCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGU- (((((...(.......).)))))......((((((((..----.((....))..................(.((.....((((((((((.....)))))))))).)).).)))))))).- ( -37.80) >DroYak_CAF1 34694 115 - 1 GCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA----GCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGU- (((((...(.......).)))))......((((((((..----.((....))..................(.((.....((((((((((.....)))))))))).)).).)))))))).- ( -37.80) >DroAna_CAF1 31384 120 - 1 GCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCAGUGAGCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCGAAAGCUGUCAUGUCCCACAGUCACCCGUA (((((.....(.((((((((......)).))).))).).((((.((....))....)))).......))))).((((..(((((((((.(....)))))))))).....))))....... ( -41.10) >consensus GCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA____GCGGAAACGGAAAUCACAACACAACGCGCGGACUAAACAUGACAGCCAAAAGCUGUCAUGUCCCACAGCAAUCCGU_ (((((...(.......).)))))......((((((((.......((....))..................(.((.....((((((((((.....)))))))))).)).).)))))))).. (-36.37 = -36.57 + 0.20)

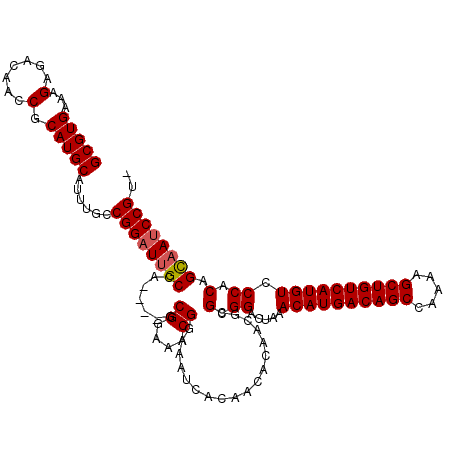

| Location | 14,789,826 – 14,789,933 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 98.88 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -25.57 |
| Energy contribution | -25.77 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14789826 107 + 23771897 UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGCUUUUUACGCACCGCACGCUACGUGUCGCCGCCAAUAAAAAGUACAAACAAAAAACCGACAGCCGGAAA- ......((((((.......(((((((...........((((((((.((..((((((...)))).))..))...))))))))..........))))).))))))))..- ( -27.71) >DroSec_CAF1 32992 107 + 1 UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGCUUUUUACGCACCGCACGCUACGUGUCGCCGCCAAUAAAAAGUACAAACAAAAAACCGACAGCCGGAAA- ......((((((.......(((((((...........((((((((.((..((((((...)))).))..))...))))))))..........))))).))))))))..- ( -27.71) >DroSim_CAF1 25243 107 + 1 UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGCUUUUUACGCACCUCACGCUACGUGUCGCCGCCAAUAAAAAGUACAAACAAAAAACCGACAGCCGGAAA- ......((((((.......(((((((...........((((((((.((.(..((((...))))..)..))...))))))))..........))))).))))))))..- ( -25.61) >DroEre_CAF1 32403 107 + 1 UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGCUUUUUACGCACCGCACGCUACGUGUCGCCGCCAAUAAAAAGUACAAACAAAAAAGCGACAGCCGGAAA- ......((((((....((.((((((.((.......))((.......)))))))).))....((((((..........................))))))))))))..- ( -30.37) >DroYak_CAF1 34770 108 + 1 UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGCUUUUUACGCACCGCACGCUACGUGUCGCCGCCAAUAAAAAGUACAAACAAAAAACCGACAGCCGGAAAA ......((((((.......(((((((...........((((((((.((..((((((...)))).))..))...))))))))..........))))).))))))))... ( -27.71) >consensus UGGCAAUCCGGCAAAUGCAUGCGGUUGUCUCUUUCACGCUUUUUACGCACCGCACGCUACGUGUCGCCGCCAAUAAAAAGUACAAACAAAAAACCGACAGCCGGAAA_ ......((((((....((.((((((.((.......))((.......)))))))).))....(((((............................)))))))))))... (-25.57 = -25.77 + 0.20)

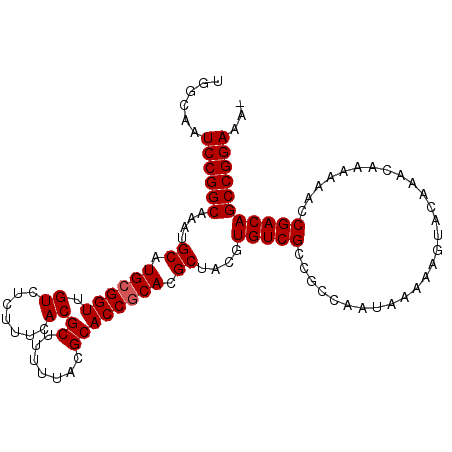

| Location | 14,789,826 – 14,789,933 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 98.88 |
| Mean single sequence MFE | -35.96 |
| Consensus MFE | -33.74 |
| Energy contribution | -33.94 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14789826 107 - 23771897 -UUUCCGGCUGUCGGUUUUUUGUUUGUACUUUUUAUUGGCGGCGACACGUAGCGUGCGGUGCGUAAAAAGCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA -..((((((((((((((((((.((((..((((((((..(((.((.((((...)))))).)))))))))))..)))).)))).)))))...)))...))))))...... ( -36.00) >DroSec_CAF1 32992 107 - 1 -UUUCCGGCUGUCGGUUUUUUGUUUGUACUUUUUAUUGGCGGCGACACGUAGCGUGCGGUGCGUAAAAAGCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA -..((((((((((((((((((.((((..((((((((..(((.((.((((...)))))).)))))))))))..)))).)))).)))))...)))...))))))...... ( -36.00) >DroSim_CAF1 25243 107 - 1 -UUUCCGGCUGUCGGUUUUUUGUUUGUACUUUUUAUUGGCGGCGACACGUAGCGUGAGGUGCGUAAAAAGCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA -..((((((((((((((((((.((((..((((((......(....)(((((.(....).)))))))))))..)))).)))).)))))...)))...))))))...... ( -33.10) >DroEre_CAF1 32403 107 - 1 -UUUCCGGCUGUCGCUUUUUUGUUUGUACUUUUUAUUGGCGGCGACACGUAGCGUGCGGUGCGUAAAAAGCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA -..(((((((((((((.....((..(((.....)))..)))))))))....(((((((((((((.....))))..........)))))))))....))))))...... ( -38.70) >DroYak_CAF1 34770 108 - 1 UUUUCCGGCUGUCGGUUUUUUGUUUGUACUUUUUAUUGGCGGCGACACGUAGCGUGCGGUGCGUAAAAAGCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA ...((((((((((((((((((.((((..((((((((..(((.((.((((...)))))).)))))))))))..)))).)))).)))))...)))...))))))...... ( -36.00) >consensus _UUUCCGGCUGUCGGUUUUUUGUUUGUACUUUUUAUUGGCGGCGACACGUAGCGUGCGGUGCGUAAAAAGCGUGAAAGAGACAACCGCAUGCAUUUGCCGGAUUGCCA ...(((((((((((.....((((..(((.....)))..)))))))))....(((((((((((((.....))))..........)))))))))....))))))...... (-33.74 = -33.94 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:54 2006