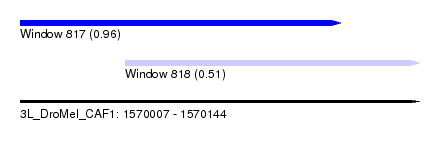
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,570,007 – 1,570,144 |
| Length | 137 |
| Max. P | 0.956867 |

| Location | 1,570,007 – 1,570,117 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 64.90 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -8.72 |
| Energy contribution | -9.04 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
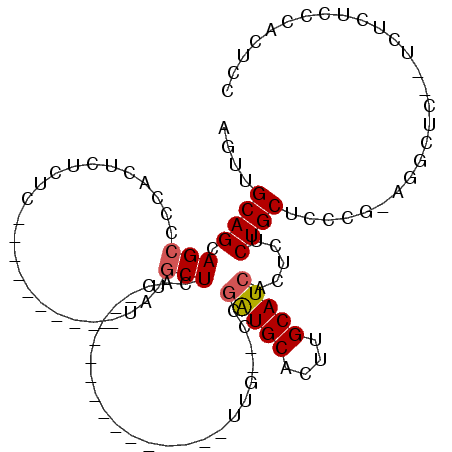
>3L_DroMel_CAF1 1570007 110 + 23771897 AGUUGCAGCAGCUCU-CUCUCUCUC---UUUUUUAUGCUCACUUCCUUGCUCCCAAGUUGUGCAAGUGCAUCGGCAUAACUUUCUGCCUCCUCAGGUUCUCUCUCUCCCAGUUC (((((...)))))..-.........---....(((((((......(((((...........)))))......)))))))......((((....))))................. ( -15.00) >DroSim_CAF1 14722 102 + 1 AGUGGCAGCAGCUCCACUCUCUCUCUGCCUUAAUAUGCUAACUUCCUCACUCGCAAGUUG--CCGAUGCACUUGCAUCACUCUCUGCUUCC----------UCUCUCCCACUCC ((.((.(((((.........................((.(((((.(......).))))))--).(((((....))))).....))))).))----------.)).......... ( -20.50) >DroEre_CAF1 20689 85 + 1 AGUUGCAGCAGACUCACUCUCUU----------UAUGCUCUC---------------UUG--CCGAUGCACUUGCAUCACUCUCUGCUCACGGAGGCUC--GCUCUCCCACUCC .((.(.((((((...........----------...((....---------------..)--).(((((....)))))....)))))))))(((((...--.).))))...... ( -21.80) >DroYak_CAF1 15030 85 + 1 AGUUGCAGCAGCCCCACUCUCUU----------UACGCUCUC---------------UUG--CCGAUGCACUUGCAUCACUCUCUGCUCCCGUAGGCUC--ACUCUCCCACUUC ..(((((((((............----------...((....---------------..)--).(((((....))))).....)))))...))))....--............. ( -14.80) >consensus AGUUGCAGCAGCCCCACUCUCUC__________UAUGCUCAC_______________UUG__CCGAUGCACUUGCAUCACUCUCUGCUCCCG_AGGCUC__UCUCUCCCACUCC ....((((.(((........................))).........................(((((....))))).....))))........................... ( -8.72 = -9.04 + 0.31)

| Location | 1,570,043 – 1,570,144 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 71.36 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -9.49 |
| Energy contribution | -9.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.49 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
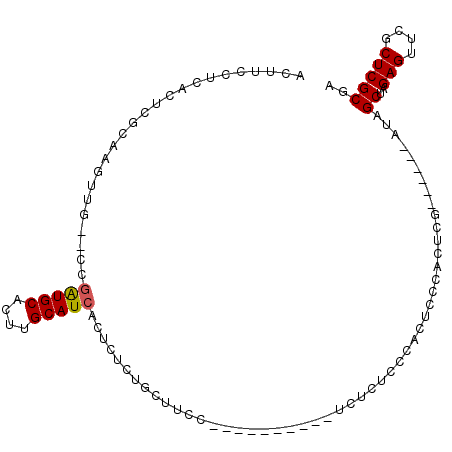
>3L_DroMel_CAF1 1570043 101 + 23771897 ACUUCCUUGCUCCCAAGUUGUGCAAGUGCAUCGGCAUAACUUUCUGCCUCCUCAGGUUCUCUCUCUCCCAGUUCCACUCG------AUAGCUAGAGUUCGCUCGCGA .....(((((...........)))))((((((((((........))))......((..((.........))..))....)------)).....(((....)))))). ( -18.40) >DroSec_CAF1 14509 89 + 1 ACCUCCUCACUCGCAAGUUG--CCGAUGCACUUGCAUCACUCUCUGCUUCC----------UCUCUCCCACUCCCACUCG------AUAGCUAGAGUUCGCUCGCGA ..........((((.(((..--..(((((....)))))(((((..(((...----------((................)------).))).)))))..))).)))) ( -19.29) >DroSim_CAF1 14762 89 + 1 ACUUCCUCACUCGCAAGUUG--CCGAUGCACUUGCAUCACUCUCUGCUUCC----------UCUCUCCCACUCCCACUCG------AUAGCUAGAGUUCGCUCGCGA ..........((((.(((..--..(((((....)))))(((((..(((...----------((................)------).))).)))))..))).)))) ( -19.29) >DroYak_CAF1 15060 88 + 1 UC---------------UUG--CCGAUGCACUUGCAUCACUCUCUGCUCCCGUAGGCUC--ACUCUCCCACUUCCACUCCUACUCAAUGGCUAGAGCACGCUCGCGA ((---------------(.(--(((((((....))))).............(((((...--.................))))).....))).)))((......)).. ( -19.75) >consensus ACUUCCUCACUCGCAAGUUG__CCGAUGCACUUGCAUCACUCUCUGCUUCC__________UCUCUCCCACUCCCACUCG______AUAGCUAGAGUUCGCUCGCGA ........................(((((....)))))...................................................((..(((....))))).. ( -9.49 = -9.55 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:43 2006