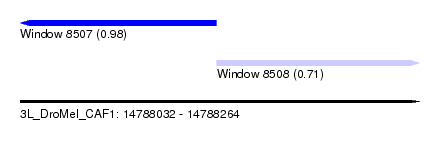
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,788,032 – 14,788,264 |
| Length | 232 |
| Max. P | 0.979640 |

| Location | 14,788,032 – 14,788,146 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -26.68 |
| Energy contribution | -26.82 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14788032 114 - 23771897 GUCCG--CUCUACGUUCACAUUUUUCUUUUUAAUAGGAAAAACACGUCUUCAGUCUC--CUGGCCAUGGAGU--GCGUGUUUGGCCAGCUUUGUUUUCGCAUGUGAAAGUAUUUAAUUCA .....--...(((.(((((((..............((..((((((((((((((((..--..)))..))))).--))))))))..)).((.........))))))))).)))......... ( -29.50) >DroSec_CAF1 31184 114 - 1 GUCCG--CUCUACGUUCACAUUUUUCUUUUUAAUAGGAAAAACACGUCUUCAGUCUC--CUGGCCAUGGAGU--GCGUGUUUGGCCAGCUUUGUUUUCGCAUGUGAAAGUAUUUAAUUCA .....--...(((.(((((((..............((..((((((((((((((((..--..)))..))))).--))))))))..)).((.........))))))))).)))......... ( -29.50) >DroSim_CAF1 23428 114 - 1 GUCCG--CUCUACGUUCACAUUUUUCUUUUUAAUAGGAAAAACACGUCUUCAGUCUC--CUGGCCAUGGAGU--GCGUGUUUGGCCAGCUUUGUUUUCGCAUGUGAAAGUAUUUAAUUCA .....--...(((.(((((((..............((..((((((((((((((((..--..)))..))))).--))))))))..)).((.........))))))))).)))......... ( -29.50) >DroEre_CAF1 30677 114 - 1 GUCCU--CUCUACGUUCACAUUUUUCUUUUUAAUAGGAAAAACACGUCUUCGGUCUC--CUGGCCAUGGAGU--GCGUGUUUGGCCAGCUUUGUUUUCGCAUGUGAAAGUAUUUAAUUCA .....--...(((.(((((((..............((..((((((((((((((((..--..))))..)))).--))))))))..)).((.........))))))))).)))......... ( -31.20) >DroYak_CAF1 33011 114 - 1 GUCCU--CUCUACGUUCACAUUUUUCUUUUUAAUAGGAAAAACACGUCUUCAGUCUC--CUGGCCAUGGAGU--GCGUGUUUGGCCAGCUUUGUUUUCGCAUGUGAAAGUAUUUAAUUCA .....--...(((.(((((((..............((..((((((((((((((((..--..)))..))))).--))))))))..)).((.........))))))))).)))......... ( -29.50) >DroAna_CAF1 30010 120 - 1 GUCCUUUCUCUCCUUUCACAUUUUUCUUUUUAAUAGGAAAAACACGUCUUCGAGCCCCACAGGCCCCGGAGUGUGCGUGUUUGGCCAGCUUUGUUUUCGCAUGUGAAAGUAUUUAAUUCA ............(((((((((..............((..(((((((((((((.(((.....)))..)))))...))))))))..)).((.........)))))))))))........... ( -33.00) >consensus GUCCG__CUCUACGUUCACAUUUUUCUUUUUAAUAGGAAAAACACGUCUUCAGUCUC__CUGGCCAUGGAGU__GCGUGUUUGGCCAGCUUUGUUUUCGCAUGUGAAAGUAUUUAAUUCA ..........(((.(((((((..............((..((((((((((((((((......)))..)))))...))))))))..)).((.........))))))))).)))......... (-26.68 = -26.82 + 0.14)

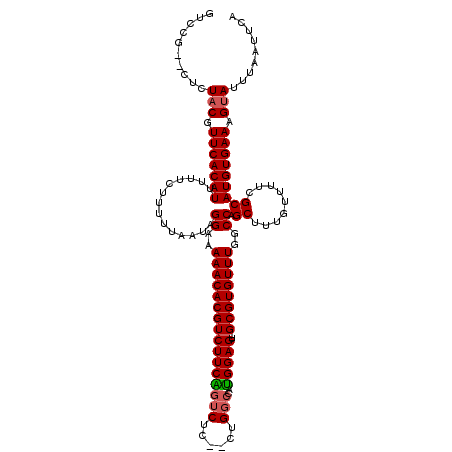
| Location | 14,788,146 – 14,788,264 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -17.73 |
| Energy contribution | -19.15 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14788146 118 + 23771897 UUGAAAGGCUGGCUCCGAAGGGAAA--CGGCCAUGAAUCUGUGCCAGAUGAAACAAUGGCAGGCACAAGCCUGGGAUUCAAUGAUAACAAAUUGAACGUGUGCUGGGAAAAUGUAUAGGG (((...(((((..(((....)))..--))))).(((((((..((((..........))))((((....)))).))))))).......)))......(.(((((.........))))).). ( -38.80) >DroSec_CAF1 31298 118 + 1 UUGAAAGGCUGGCUCCGAAGGGAAA--UGGCCAUGAAUCUGUGCCAGAUGAAACAAUGGCAGGCACAAGCCUGGGAUUCAAUGAUAACAAAUUGAACGUGUGAUGGGAAAAUUUAUAGGG (((...(((((..(((....)))..--))))).(((((((..((((..........))))((((....)))).))))))).......)))......(.(((((.........))))).). ( -33.60) >DroSim_CAF1 23542 118 + 1 UUGAAAGGCUGGCUCCGAAGGGAAA--CGGCCAUGAAUCUGUGCCAGAUGAAACAAUGGCAGGCACAAGCCUGGGAUUCAAUGAUAACAAAUUGAACGUGUGAUGGGAAAAUUUAUAGGG (((...(((((..(((....)))..--))))).(((((((..((((..........))))((((....)))).))))))).......)))......(.(((((.........))))).). ( -36.70) >DroEre_CAF1 30791 118 + 1 UUGAAAGGCUGGCUCCGAAGGGAAA--UGGCUAUGAAUCUGUGCCAGAUGAAACAAUGGCAGGCAAAAGCCUGGGAUUCAAUGAUAACAAAUUGAACGUGUGCUGGGGAAAUUUAUAGGG ......(((((..(((....)))..--)))))((((((...(.((((.....(((.((.(((((....)))))...((((((........))))))))))).)))).)..)))))).... ( -32.10) >DroYak_CAF1 33125 120 + 1 UUGAAAGGCUGACUCCAAAGGGAAACACGACUGUGAAUCUGUGCCAGAUGAAACAAUGGCAGGCAAAAGCCUGGGAUUCAAUGAUAACAAAUUGAACGUGUGCUGGGAAAAUUUAUAGGG ..............((....(....)....((((((((...(.((((.....(((.((.(((((....)))))...((((((........))))))))))).)))).)..)))))))))) ( -28.30) >DroAna_CAF1 30130 93 + 1 CUGAAAAGCU----------------------ACGAAUGUGUGUUAGAUGAAUUAAUAGCCUA-AUAUGGUCAGAAUACAAGGAUAACAAAUAGAUCGUA----GGGCAAGUGUGGAAUG ........((----------------------((((.(((.(((((..((.(((....(((..-....)))...))).))....))))).)))..)))))----)............... ( -15.70) >consensus UUGAAAGGCUGGCUCCGAAGGGAAA__CGGCCAUGAAUCUGUGCCAGAUGAAACAAUGGCAGGCACAAGCCUGGGAUUCAAUGAUAACAAAUUGAACGUGUGCUGGGAAAAUUUAUAGGG ......(((((..(((....)))....))))).(((((((..((((..........))))((((....)))).)))))))........................................ (-17.73 = -19.15 + 1.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:48 2006