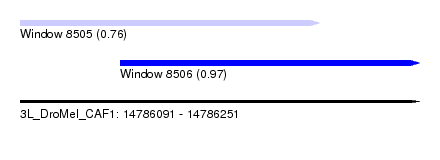
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,786,091 – 14,786,251 |
| Length | 160 |
| Max. P | 0.965158 |

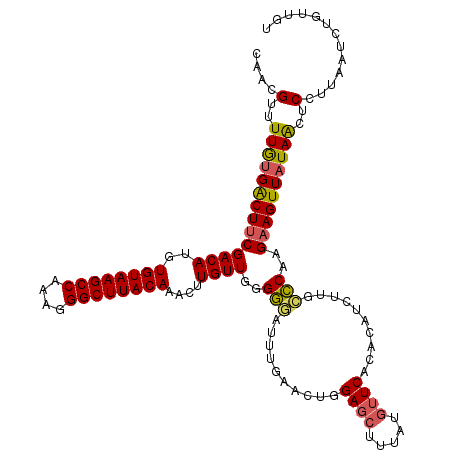
| Location | 14,786,091 – 14,786,211 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.50 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -28.76 |
| Energy contribution | -29.57 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14786091 120 + 23771897 GAUUCUGGACGUAAAGCACUGAAAACAAGAGGGAUAAUUGCAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACAUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCA ......(((((((((((.((....((((((.(...........).))))))..((((((((((((((((((....)))))))..)))))))))))..........)).))))))))))). ( -44.00) >DroSec_CAF1 29241 120 + 1 GAUUCUGGACGUAAAACCAUGAAAACAAGAGGGAUAAUUGCAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCA ......(((((((((.(((.....((((((.(...........).))))))..((((((((..((((((((....))))))))....)))))))).........)))...))))))))). ( -36.00) >DroSim_CAF1 21436 120 + 1 GAUUCUGGACGUAAAACCAUGAAAACAAGAGGGAUAAUUGCAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCA ......(((((((((.(((.....((((((.(...........).))))))..((((((((..((((((((....))))))))....)))))))).........)))...))))))))). ( -36.00) >DroEre_CAF1 28704 120 + 1 AAUACUGGACGUAAAACCCCCAAAACAAGAGGGAUAAUUGCAACGUUUUGCGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCA ......(((((((((.((((((..(((((...((...((((((....))))))..))......((((((((....))))))))..))))))))))).(((.....)))..))))))))). ( -39.70) >DroYak_CAF1 29281 120 + 1 GAUACUGGACGUAAAACCCUGAACACAAGAGGGAUAAUUGCAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUCAACUGGAGCUUUAUGUUCA ......(((((((((.((((((((((((((.(...........).))))))).((((((((..((((((((....))))))))....))))))))...))))...)).).))))))))). ( -40.60) >DroAna_CAF1 28069 115 + 1 GCUUUCAAAUCGAAUUCCUCCGGAAAGGGAUGGAUAAUUGCUGCGGUUUAUGGCUCCGACAUGUGUAAGCCAAAGGGCUUACAAAGUUGUUGGGUAUUU-----UCGAGGUUUAUGGUCC .............((((((......))))))((((....(((.(((......((.((((((..((((((((....))))))))....))))))))....-----))).))).....)))) ( -34.30) >consensus GAUUCUGGACGUAAAACCCUGAAAACAAGAGGGAUAAUUGCAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCA ......(((((((((.........((((((.(...........).))))))..((((((((..((((((((....))))))))....))))))))...............))))))))). (-28.76 = -29.57 + 0.81)

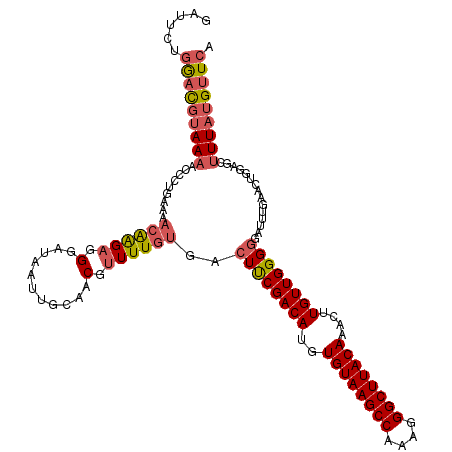
| Location | 14,786,131 – 14,786,251 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -30.58 |
| Energy contribution | -30.50 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14786131 120 + 23771897 CAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACAUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCACACAUCUUGCCCAAGAAGUUAUAACUCCUUAAUCUGUUGU ....(..((((((((((((((((((((((((....)))))))..)))))))(((.((..((...((((((.....)))).))))..)).)))..))))))))))..)............. ( -39.10) >DroSec_CAF1 29281 120 + 1 CAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCACACAUCUUGCCCAAGAAGUUAUAACUCCUUAAUCUGUUGU ....(..((((((((((((((..((((((((....))))))))....))))(((.((..((...((((((.....)))).))))..)).)))..))))))))))..)............. ( -36.50) >DroSim_CAF1 21476 120 + 1 CAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCACACAUCUUGCCCAAGAAGUUAUAACUCCUUAAUCUGUUGU ....(..((((((((((((((..((((((((....))))))))....))))(((.((..((...((((((.....)))).))))..)).)))..))))))))))..)............. ( -36.50) >DroEre_CAF1 28744 120 + 1 CAACGUUUUGCGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCACACAUCUUGGCCCAGAAGUUAUAGCUCCUUAAUCUGUUGU .........(((((..(((((..((((((((....))))))))....)))))((((...(((.((((.(((..((((....))))...)))))))...)))...)))).......))))) ( -37.20) >DroYak_CAF1 29321 120 + 1 CAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUCAACUGGAGCUUUAUGUUCACACAUCUUGCCCAAGAAGUUAUAACUCCUUAAUCUGUUGU (((((..(((.((((((((((..((((((((....))))))))....))))))))((((((...(((.((...((((....))))...))))).)))))).....))..)))..))))). ( -34.90) >DroAna_CAF1 28109 115 + 1 CUGCGGUUUAUGGCUCCGACAUGUGUAAGCCAAAGGGCUUACAAAGUUGUUGGGUAUUU-----UCGAGGUUUAUGGUCCCACAUCUUGUACUAGAAGUUAUAACUCCUUAAUCCGUUGU ..((((.(((.((.(((((((..((((((((....))))))))....))))))).....-----.((((((...((....)).)))))).................)).))).))))... ( -31.50) >consensus CAACGUUUUGUGACUUCGACAUGUGUAAGCCAAAGGGCUUACAAACUUGUUGGGGGAUUUGAACUGGAGCUUUAUGUUCACACAUCUUGCCCAAGAAGUUAUAACUCCUUAAUCUGUUGU ....(..((((((((((((((..((((((((....))))))))....))))..(((..........((((.....))))..........)))..))))))))))..)............. (-30.58 = -30.50 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:46 2006