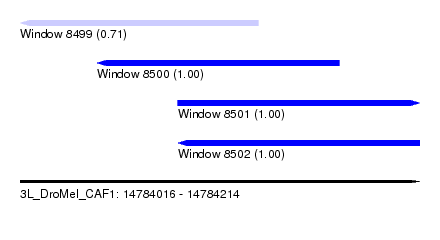
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,784,016 – 14,784,214 |
| Length | 198 |
| Max. P | 0.999849 |

| Location | 14,784,016 – 14,784,134 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.56 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -26.77 |
| Energy contribution | -26.66 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14784016 118 - 23771897 CUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCUGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGAUUUAUAUUGCUCACAAACAUAUGGAAAAUAUAUA--UACU .((((.((((((((.(((....)))))))))))..))))..((((((((...............))))).))).........((((((..((.((......)))).))))))..--.... ( -30.76) >DroEre_CAF1 26298 113 - 1 CUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCUGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGAUUUAUAUAGCUCACAAACUUAUGGAAACUAUAU------- .((((.((((((((.(((....)))))))))))..))))....((((((...............))))))(((.(((((.....))))).))).........(....).....------- ( -31.06) >DroYak_CAF1 27234 119 - 1 CUGGCAGGCGCAAGUGUUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCUGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGAUUUAUAUUGCUCACAAACAUAUGGAAAAUAUAUACG-AGU .((((.((((((((.((.....)).))))))))..)))).(((((((((...............))))))............((((((..((.((......)))).))))))...)-)). ( -28.86) >consensus CUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCUGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGAUUUAUAUUGCUCACAAACAUAUGGAAAAUAUAUA___A_U .((((.((((((((.(((....)))))))))))..))))..((((((((...............))))).))).......((((((.((........))))))))............... (-26.77 = -26.66 + -0.11)


| Location | 14,784,054 – 14,784,174 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.15 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -29.72 |
| Energy contribution | -29.92 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14784054 120 - 23771897 CACUUAUAUAUAUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCUGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGA ....................................(((((.(((.((((((((.(((....))))))))))).((((......))))((....))..........)))..))))).... ( -32.00) >DroSec_CAF1 27063 118 - 1 CACUUAUAU--AUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCAGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGA .........--.........................(((((.(((.((((((((.(((....)))))))))))..(((......))).((....))..........)))..))))).... ( -31.50) >DroSim_CAF1 19270 118 - 1 CACUUAUAU--AUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCAGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGA .........--.........................(((((.(((.((((((((.(((....)))))))))))..(((......))).((....))..........)))..))))).... ( -31.50) >DroEre_CAF1 26331 118 - 1 CACUUAUAU--AUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCUGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGA .........--.........................(((((.(((.((((((((.(((....))))))))))).((((......))))((....))..........)))..))))).... ( -32.00) >DroYak_CAF1 27273 118 - 1 CACUUAUAU--AUAUGAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGUUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCUGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGA .........--.........................(((((.(((.((((((((.((.....)).)))))))).((((......))))((....))..........)))..))))).... ( -28.70) >consensus CACUUAUAU__AUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAUCUCAGGCUGCAAAUGCAUUAAAUCCAGCCUUGAACUAUGA ....................................(((((.(((.((((((((.(((....)))))))))))..(((......))).((....))..........)))..))))).... (-29.72 = -29.92 + 0.20)

| Location | 14,784,094 – 14,784,214 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.18 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14784094 120 + 23771897 AUGGCUUAGCACAAGGGCGAAAACCACUUGCGCCUGCCAGAACUGGAAUAAAAAUGCAAUGUUUUAUAUAUAUAUAAGUGUAUCCACAAACAAACGUGUAUAAAAUGCGCAUAUAUAACA .((((...((.((((((......)).)))).))..))))..............((((...(((((((((((.......(((....))).......)))))))))))..))))........ ( -26.74) >DroSec_CAF1 27103 118 + 1 AUGGCUUAGCACAAGGGCGAAAACCACUUGCGCCUGCCAGAACUGGAAUAAAAAUGCAAUGUUUUAUAU--AUAUAAGUGCAUCCACAAACAAACGUGUAAUAAAUGCGCAUAUAUAACA .((((...((.((((((......)).)))).))..))))........((((((........)))))).(--(((((.((((((.(((........)))......)))))).))))))... ( -26.40) >DroSim_CAF1 19310 118 + 1 AUGGCUUAGCACAAGGGCGAAAACCACUUGCGCCUGCCAGAACUGGAAUAAAAAUGCAAUGUUUUAUAU--AUAUAAGUGCAUCCACAAACAAACGUGUAUAAAAUGCGCAUAUAUAACA .((((...((.((((((......)).)))).))..))))........((((((........)))))).(--(((((.((((((.(((........)))......)))))).))))))... ( -26.40) >DroEre_CAF1 26371 118 + 1 AUGGCUUAGCACAAGGGCGAAAACCACUUGCGCCUGCCAGAACUGGAAUAAAAAUGCAAUGUUUUAUAU--AUAUAAGUGCAUUCGCAAGCAAACGUAUAUAAAAUGCGCAUAUAUAACA .((((...((.((((((......)).)))).))..))))..............((((...(((((((((--((.....(((........)))...)))))))))))..))))........ ( -28.70) >DroYak_CAF1 27313 118 + 1 AUGGCUUAGCACAAGGGCGAAAAACACUUGCGCCUGCCAGAACUGGAAUAAAAAUGCAAUGUUUCAUAU--AUAUAAGUGCAUCCACAAACAAAUGUAUAUGAAAUGCGCAUAUAUAACA .((((...((.(((((........).)))).))..))))..............((((...(((((((((--((((..((..........))..)))))))))))))..))))........ ( -28.20) >consensus AUGGCUUAGCACAAGGGCGAAAACCACUUGCGCCUGCCAGAACUGGAAUAAAAAUGCAAUGUUUUAUAU__AUAUAAGUGCAUCCACAAACAAACGUGUAUAAAAUGCGCAUAUAUAACA .((((...((.((((((......)).)))).))..))))....(((((((.........))))))).....(((((.((((((......((....)).......)))))).))))).... (-23.46 = -23.18 + -0.28)


| Location | 14,784,094 – 14,784,214 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -32.46 |
| Consensus MFE | -29.06 |
| Energy contribution | -28.94 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14784094 120 - 23771897 UGUUAUAUAUGCGCAUUUUAUACACGUUUGUUUGUGGAUACACUUAUAUAUAUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAU ((..(((.(((((..(((((((...((.(((..(((....)))..))).)).)))))))...)))))...)))..))....((((.((((((((.(((....)))))))))))..)))). ( -31.90) >DroSec_CAF1 27103 118 - 1 UGUUAUAUAUGCGCAUUUAUUACACGUUUGUUUGUGGAUGCACUUAUAU--AUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAU ((..(((.((((((((((((.(((....)))..))))))))..((((..--..))))......))))...)))..))....((((.((((((((.(((....)))))))))))..)))). ( -33.50) >DroSim_CAF1 19310 118 - 1 UGUUAUAUAUGCGCAUUUUAUACACGUUUGUUUGUGGAUGCACUUAUAU--AUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAU ((..(((.(((((((((.....((((......)))))))))..((((..--..))))......))))...)))..))....((((.((((((((.(((....)))))))))))..)))). ( -31.30) >DroEre_CAF1 26371 118 - 1 UGUUAUAUAUGCGCAUUUUAUAUACGUUUGCUUGCGAAUGCACUUAUAU--AUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAU ((..(((.(((((..(((((((((..(.(((........)))...)..)--))))))))...)))))...)))..))....((((.((((((((.(((....)))))))))))..)))). ( -33.70) >DroYak_CAF1 27313 118 - 1 UGUUAUAUAUGCGCAUUUCAUAUACAUUUGUUUGUGGAUGCACUUAUAU--AUAUGAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGUUUUUCGCCCUUGUGCUAAGCCAU ((..(((.(((((..(((((((((....(((..(((....)))..))))--))))))))...)))))...)))..))....((((.((((((((.((.....)).))))))))..)))). ( -31.90) >consensus UGUUAUAUAUGCGCAUUUUAUACACGUUUGUUUGUGGAUGCACUUAUAU__AUAUAAAACAUUGCAUUUUUAUUCCAGUUCUGGCAGGCGCAAGUGGUUUUCGCCCUUGUGCUAAGCCAU ((..(((.(((((((((.....((((......)))))))))..(((((....)))))......))))...)))..))....((((.((((((((.(((....)))))))))))..)))). (-29.06 = -28.94 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:42 2006