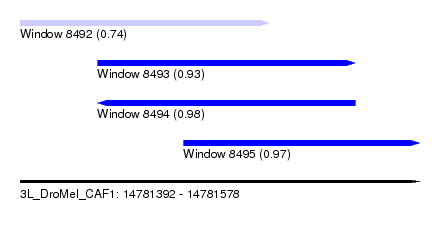
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,781,392 – 14,781,578 |
| Length | 186 |
| Max. P | 0.975315 |

| Location | 14,781,392 – 14,781,508 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.26 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -32.18 |
| Energy contribution | -32.73 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
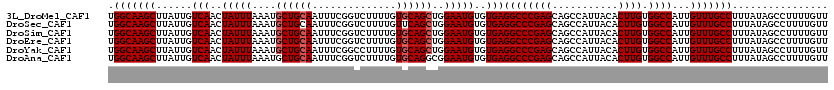
>3L_DroMel_CAF1 14781392 116 + 23771897 UUGACGCCCCCACACAG----CAGCUGGCCAAAAACUCGUUGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCA .....(((..((((((.----((((((..(((((....((((((((.....)))))))).........((((......)))).)))))..))))))...)))))).)))........... ( -36.50) >DroSec_CAF1 24332 120 + 1 UUGACGCCCCCACACAGCAAGCAGCUGGCCAAAAACUCGUUGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUUCAGCUGGAAUGUGUGAGGCCCGAGCAGCCA .....(((..((((((.....((((((..(((((....((((((((.....)))))))).........((((......)))).)))))..))))))...)))))).)))........... ( -37.90) >DroSim_CAF1 16505 120 + 1 UUGACGCCCCCACACAGCAAGCAGCUGGCCAAAAACUCGUUGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCA .....(((..((((((.....((((((..(((((....((((((((.....)))))))).........((((......)))).)))))..))))))...)))))).)))........... ( -35.90) >DroEre_CAF1 23618 114 + 1 UUGACGCC--CACACAG----CAGCUGGCCAAAAACUCGUUGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCA .....(((--((((((.----((((((..(((((....((((((((.....)))))))).........((((......)))).)))))..))))))...)))))).)))........... ( -37.70) >DroYak_CAF1 24554 116 + 1 UUGACGCCCCCACACAG----CAGCUGGCCAAAAACUCGUUGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGCCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCA .....(((..((((((.----((((((..(((((....((((((((.....)))))))).........((((......)))).)))))..))))))...)))))).)))........... ( -38.40) >DroAna_CAF1 24036 113 + 1 UCACCACCCCCG--CAG----CAGCUGGCCAA-AACUCGUUGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGGCGGAAUGUGUGAGGCCCGAGCAGCCA ((((((...(((--(.(----(((..((((..-.....((((((((.....)))))))).....((((......)))).))))..))))....))))..)).))))(((.......))). ( -33.50) >consensus UUGACGCCCCCACACAG____CAGCUGGCCAAAAACUCGUUGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCA ..........((((((.....((((((..(((((....((((((((.....)))))))).........((((......)))).)))))..))))))...)))))).(((.......))). (-32.18 = -32.73 + 0.56)

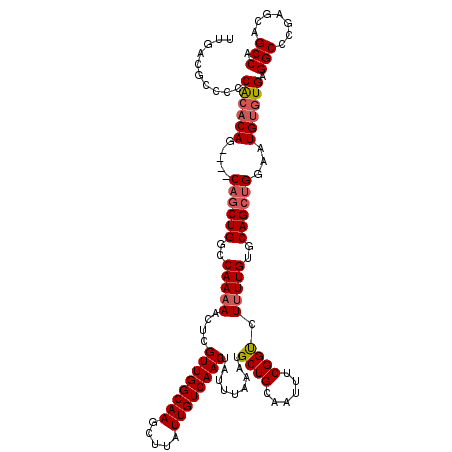
| Location | 14,781,428 – 14,781,548 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -30.96 |
| Energy contribution | -31.29 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14781428 120 + 23771897 UGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUU .(((((((......(((..(((((....((((((..............))))))..)))))..)))((((((((...........)))).))))...)))))))................ ( -33.04) >DroSec_CAF1 24372 120 + 1 UGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUUCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUU .(((((((..((.((((...........(((((....((((((((.(((((.....)))))...)))).)))))))))...........)))).)).)))))))................ ( -31.25) >DroSim_CAF1 16545 120 + 1 UGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUU .(((((((......(((..(((((....((((((..............))))))..)))))..)))((((((((...........)))).))))...)))))))................ ( -33.04) >DroEre_CAF1 23652 120 + 1 UGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUU .(((((((......(((..(((((....((((((..............))))))..)))))..)))((((((((...........)))).))))...)))))))................ ( -33.04) >DroYak_CAF1 24590 120 + 1 UGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGCCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUU .(((((((..((.((((...........(((((....((((((((..(..((.......))..))))).)))))))))...........)))).)).)))))))................ ( -33.45) >DroAna_CAF1 24069 120 + 1 UGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGGCGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUU .(((((((...................(((.((((..........)))))))(((.(...(((((((((.......))).))))))...).)))...)))))))................ ( -32.00) >consensus UGGCAAGCUUAUUGUCAACUAUUUAAAUGCUGCAAUUUCGGUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUU .(((((((......(((..(((((....((((((..............))))))..)))))..)))((((((((...........)))).))))...)))))))................ (-30.96 = -31.29 + 0.33)

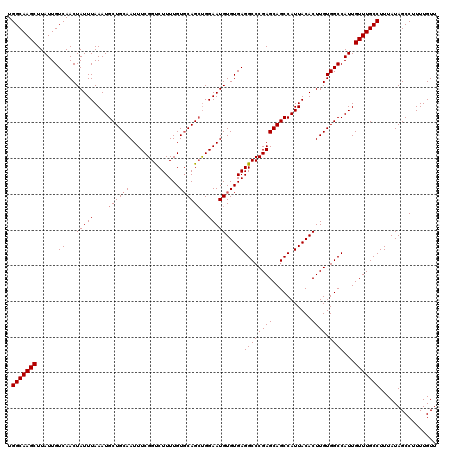
| Location | 14,781,428 – 14,781,548 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14781428 120 - 23771897 AACAAAAGGCUAUAAAGGCAAACAAUGGCCACAAGUGUAAUGGCUGCUCGGGCCUCACACAUUCCAGCUGCACAAAAGACCGAAAUUGCAGCAUUUAAAUAGUUGACAAUAAGCUUGCCA .......(((......(((........))).(((((((...((((.....))))..))))......((((((..............))))))..........)))...........))). ( -28.04) >DroSec_CAF1 24372 120 - 1 AACAAAAGGCUAUAAAGGCAAACAAUGGCCACAAGUGUAAUGGCUGCUCGGGCCUCACACAUUCCAGCUGAACAAAAGACCGAAAUUGCAGCAUUUAAAUAGUUGACAAUAAGCUUGCCA .......(((......(((........)))....((((...((((.....))))..)))).....((((...(....)......((((((((.........)))).)))).)))).))). ( -24.20) >DroSim_CAF1 16545 120 - 1 AACAAAAGGCUAUAAAGGCAAACAAUGGCCACAAGUGUAAUGGCUGCUCGGGCCUCACACAUUCCAGCUGCACAAAAGACCGAAAUUGCAGCAUUUAAAUAGUUGACAAUAAGCUUGCCA .......(((......(((........))).(((((((...((((.....))))..))))......((((((..............))))))..........)))...........))). ( -28.04) >DroEre_CAF1 23652 120 - 1 AACAAAAGGCUAUAAAGGCAAACAAUGGCCACAAGUGUAAUGGCUGCUCGGGCCUCACACAUUCCAGCUGCACAAAAGACCGAAAUUGCAGCAUUUAAAUAGUUGACAAUAAGCUUGCCA .......(((......(((........))).(((((((...((((.....))))..))))......((((((..............))))))..........)))...........))). ( -28.04) >DroYak_CAF1 24590 120 - 1 AACAAAAGGCUAUAAAGGCAAACAAUGGCCACAAGUGUAAUGGCUGCUCGGGCCUCACACAUUCCAGCUGCACAAAAGGCCGAAAUUGCAGCAUUUAAAUAGUUGACAAUAAGCUUGCCA ................(((((.(..(((((....((((...(((((....(........)....)))))))))....)))))..((((((((.........)))).))))..).))))). ( -29.90) >DroAna_CAF1 24069 120 - 1 AACAAAAGGCUAUAAAGGCAAACAAUGGCCACAAGUGUAAUGGCUGCUCGGGCCUCACACAUUCCGCCUGCACAAAAGACCGAAAUUGCAGCAUUUAAAUAGUUGACAAUAAGCUUGCCA .......(((.....((((......((....)).((((...((((.....))))..)))).....))))...............((((((((.........)))).))))......))). ( -25.50) >consensus AACAAAAGGCUAUAAAGGCAAACAAUGGCCACAAGUGUAAUGGCUGCUCGGGCCUCACACAUUCCAGCUGCACAAAAGACCGAAAUUGCAGCAUUUAAAUAGUUGACAAUAAGCUUGCCA .......(((......(((........))).(((((((...((((.....))))..))))......((((((..............))))))..........)))...........))). (-25.82 = -26.16 + 0.33)

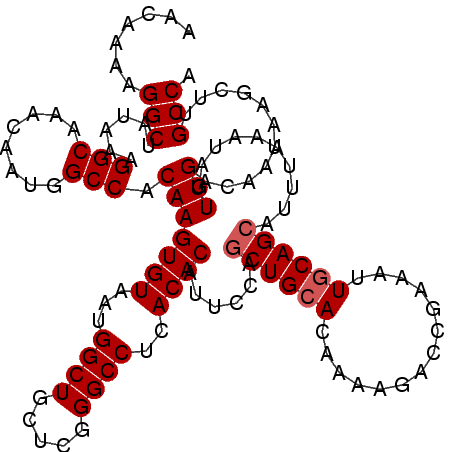

| Location | 14,781,468 – 14,781,578 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.87 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -27.45 |
| Energy contribution | -27.78 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.970853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14781468 110 + 23771897 GUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUUGCAUAAAUCGUUUACUACAAUAAAA----AAAAA------ ....((((((((((.((..((((.(((((..((((((((((.......)))))...))))))))))))))..))...))))))))))..................----.....------ ( -29.50) >DroSec_CAF1 24412 104 + 1 GUCUUUUGUUCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUUGCAUAAAUCGUUUACUACAAUAAA---------------- ....(((((.((((.((..((((.(((((..((((((((((.......)))))...))))))))))))))..))...)))).))))).................---------------- ( -22.90) >DroSim_CAF1 16585 104 + 1 GUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUUGCAUAAAUCGUUUACUACAAUAAA---------------- ....((((((((((.((..((((.(((((..((((((((((.......)))))...))))))))))))))..))...)))))))))).................---------------- ( -29.50) >DroEre_CAF1 23692 116 + 1 GUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUUGCAUAAAUCGUUUACUACAAUAAAA----AAAAAAACAAA ....((((((((((.((..((((.(((((..((((((((((.......)))))...))))))))))))))..))...))))))))))..................----........... ( -29.50) >DroYak_CAF1 24630 106 + 1 GCCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUUGCAUAAAUCGUUUACUACAAUAAAA----A---------- ((..((((((((((.((..((((.(((((..((((((((((.......)))))...))))))))))))))..))...))))))))))..))..............----.---------- ( -30.20) >DroAna_CAF1 24109 114 + 1 GUCUUUUGUGCAGGCGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUUGCAUAAAUCGUUUACAACAAUACAGCAGCAGAAA------ ...((((((((.(((.(...(((((((((.......))).))))))...).))).((((..((.(((((.((........)))))))..))..)))).......))).))))).------ ( -29.40) >consensus GUCUUUUGUGCAGCUGGAAUGUGUGAGGCCCGAGCAGCCAUUACACUUGUGGCCAUUGUUUGCCUUUAUAGCCUUUUGUUGCAUAAAUCGUUUACUACAAUAAAA____A__________ ....((((((((((.((..((((.(((((..((((((((((.......)))))...))))))))))))))..))...))))))))))................................. (-27.45 = -27.78 + 0.33)


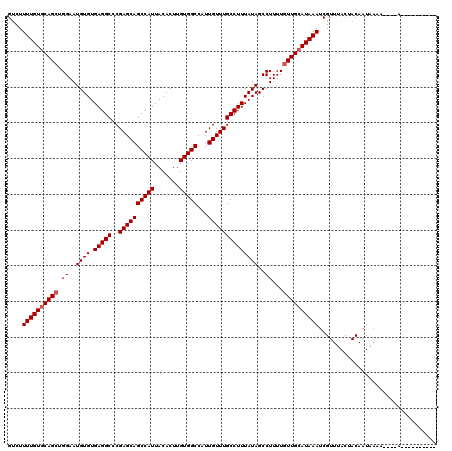
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:35 2006