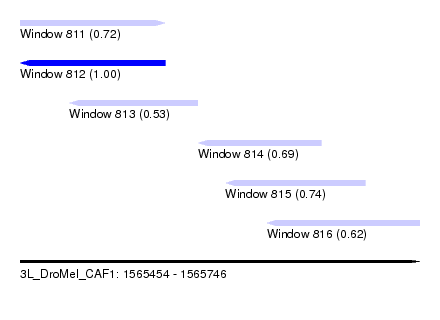
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,565,454 – 1,565,746 |
| Length | 292 |
| Max. P | 0.999495 |

| Location | 1,565,454 – 1,565,560 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -11.50 |
| Energy contribution | -16.50 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1565454 106 + 23771897 G---GCUUUUCUUUUCUUGU-UUUCCCCCAUUUCUAUCAGUCGAAUGCCGCCAUUUGGCACUCGACUGACUUUCUUUCCACUUCGGU-UUGGUCAUGGAAAUGGAAAUUUG .---................-......(((((((((((((((((.(((((.....))))).))))))))........((((....))-..))...)))))))))....... ( -32.50) >DroSec_CAF1 9936 106 + 1 GCCCGCUUUUCU-----UUUCUUUUCCCCAUUUCUGUCAGUGGAAUGCCGCCAUUUGGCACUCGACUGACUUUCUUUCCACUUCGGUUUUGGUCAUGGAAAUGGAAAUUUG ............-----..........((((((((((((((.((.(((((.....))))).)).)))))).......((((....))...))....))))))))....... ( -30.40) >DroSim_CAF1 10225 100 + 1 GCCCGCUUUUCUUUACUUUUCUUUUCCCCAUUUCUGUCAGUGGAAUGCCGCCA-----------ACUGACUUUCUUUCCACUUUGGUUUUGGUCAUGGAAAUGGAAAUUUG ...........................((((((((((..((((....))))..-----------))(((((......((.....))....))))).))))))))....... ( -20.50) >DroEre_CAF1 16076 90 + 1 CCCAGCUUUUCUUU--------UUUCCCCAUUUCGGUCACUGGAAUUCCGCCACUUGGCUC------GACUUUCUUUCCACUUCGGUU-UGGGC------AGGGAAAUUUG ..............--------((((((......((((...((....))(((....)))..------)))).....(((((....)).-.))).------.)))))).... ( -20.50) >consensus GCCCGCUUUUCUUU___UUU_UUUUCCCCAUUUCUGUCAGUGGAAUGCCGCCAUUUGGCAC___ACUGACUUUCUUUCCACUUCGGUUUUGGUCAUGGAAAUGGAAAUUUG ...........................((((((((((((((.((.(((((.....))))).)).)))))).......(((.........)))....))))))))....... (-11.50 = -16.50 + 5.00)

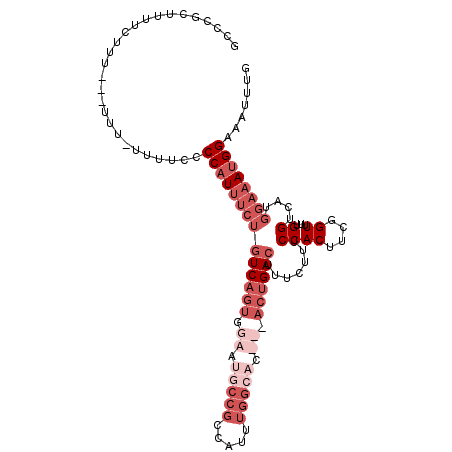
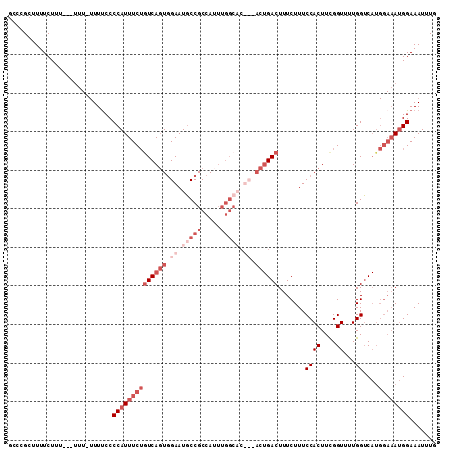
| Location | 1,565,454 – 1,565,560 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -17.40 |
| Energy contribution | -21.53 |
| Covariance contribution | 4.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1565454 106 - 23771897 CAAAUUUCCAUUUCCAUGACCAA-ACCGAAGUGGAAAGAAAGUCAGUCGAGUGCCAAAUGGCGGCAUUCGACUGAUAGAAAUGGGGGAAA-ACAAGAAAAGAAAAGC---C ....(..(((((((.....(((.-.......))).......((((((((((((((.......)))))))))))))).)))))))..)...-................---. ( -37.20) >DroSec_CAF1 9936 106 - 1 CAAAUUUCCAUUUCCAUGACCAAAACCGAAGUGGAAAGAAAGUCAGUCGAGUGCCAAAUGGCGGCAUUCCACUGACAGAAAUGGGGAAAAGAAA-----AGAAAAGCGGGC ....(..(((((((.....(((.........))).......((((((.(((((((.......))))))).)))))).)))))))..).......-----............ ( -32.70) >DroSim_CAF1 10225 100 - 1 CAAAUUUCCAUUUCCAUGACCAAAACCAAAGUGGAAAGAAAGUCAGU-----------UGGCGGCAUUCCACUGACAGAAAUGGGGAAAAGAAAAGUAAAGAAAAGCGGGC ....(..(((((((..((........)).(((((((.(...(((...-----------.)))..).)))))))....)))))))..)........................ ( -22.30) >DroEre_CAF1 16076 90 - 1 CAAAUUUCCCU------GCCCA-AACCGAAGUGGAAAGAAAGUC------GAGCCAAGUGGCGGAAUUCCAGUGACCGAAAUGGGGAAA--------AAAGAAAAGCUGGG ....(((((((------..(((-........))).......(((------(.(((....)))((....))..))))......)))))))--------.............. ( -20.30) >consensus CAAAUUUCCAUUUCCAUGACCAAAACCGAAGUGGAAAGAAAGUCAGU___GUGCCAAAUGGCGGCAUUCCACUGACAGAAAUGGGGAAAA_AAA___AAAGAAAAGCGGGC ....(..(((((((.....(((.........))).......((((((.(((((((.......))))))).)))))).)))))))..)........................ (-17.40 = -21.53 + 4.13)

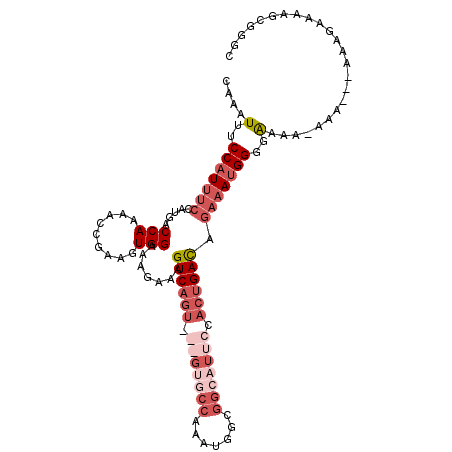
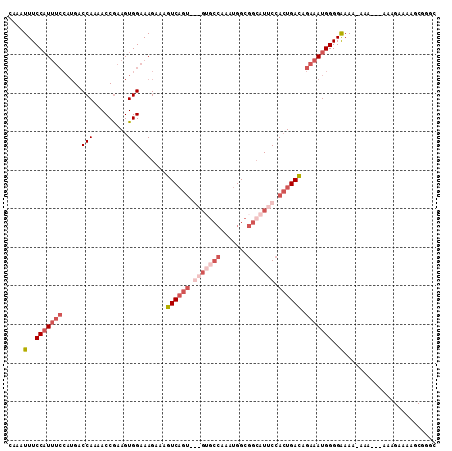
| Location | 1,565,490 – 1,565,584 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -9.12 |
| Energy contribution | -11.72 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.39 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1565490 94 - 23771897 GGUCAAAUAAG--GCGGCGAAGUUGACAAAUUUCCAUUUCCAUGACCAA-ACCGAAGUGGAAAGAAAGUCAGUCGAGUGCCAAAUGGCGGCAUUCGA .(((......)--)).......(((((...((((((((((.........-...))))))))))....)))))(((((((((.......))))))))) ( -29.50) >DroSec_CAF1 9971 94 - 1 GGUCAAAUACA--ACGGCC-AGUUGACAAAUUUCCAUUUCCAUGACCAAAACCGAAGUGGAAAGAAAGUCAGUCGAGUGCCAAAUGGCGGCAUUCCA ((((.......--..))))-.((((((...((((((((((.............))))))))))....))))).)(((((((.......))))))).. ( -28.62) >DroSim_CAF1 10265 83 - 1 GGUCAAAUACA--ACAGCC-AGUUGACAAAUUUCCAUUUCCAUGACCAAAACCAAAGUGGAAAGAAAGUCAGU-----------UGGCGGCAUUCCA ...........--...(((-(..((((...(((((((((...((........)))))))))))....))))..-----------))))((....)). ( -21.70) >DroEre_CAF1 16108 80 - 1 GGUCAAAUGA---GCGGCG-AGUUGACAAAUUUCCCU------GCCCA-AACCGAAGUGGAAAGAAAGUC------GAGCCAAGUGGCGGAAUUCCA .((((..((.---((..((-(.((......(((((((------.....-......)).)))))..)).))------).))))..))))((....)). ( -19.20) >DroYak_CAF1 10180 83 - 1 GGUCAAAUAAAGCGCGGCG-AGUUGACAAAUUUCCAU------GCCCA-AACCGAAGUGGAAAGAAAGUG------GAGCCAAGUGGCGGAAUUCCA .(((((.....((...)).-..)))))...(((((((------..(..-....)..))))))).......------..(((....)))((....)). ( -18.10) >consensus GGUCAAAUAAA__GCGGCG_AGUUGACAAAUUUCCAUUUCCAUGACCAAAACCGAAGUGGAAAGAAAGUCAGU___GAGCCAAAUGGCGGCAUUCCA .(((...................((((...(((((((((...............)))))))))....)))).......(((....))))))...... ( -9.12 = -11.72 + 2.60)

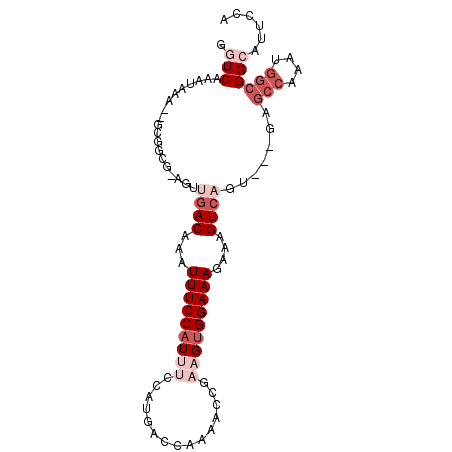
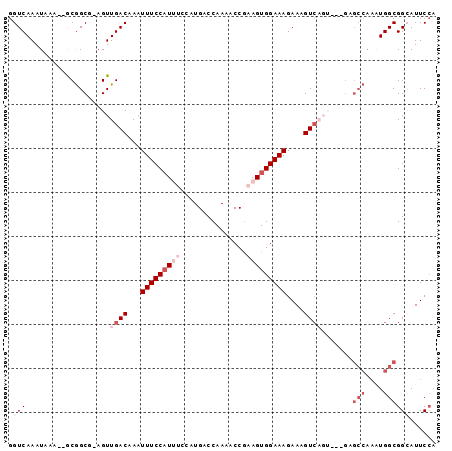
| Location | 1,565,584 – 1,565,674 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1565584 90 - 23771897 UCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAAGCG-------------------AUCGCCCAGGGCGACAAGCG------ ....(((((((((((((((((......)))))))))))))......(((((-----...(((((........)))-------------------)).))....)))))))....------ ( -24.40) >DroSec_CAF1 10065 90 - 1 UCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAAGCG-------------------AUCGCCCAGGACGACAAGCG------ ....(((((((((((((((((......)))))))))))))......(((..-----.(.(((((........)))-------------------)).)....)).)))))....------ ( -21.50) >DroSim_CAF1 10348 90 - 1 UCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAAGCG-------------------AUCGCCCAGGGCGACAAGCG------ ....(((((((((((((((((......)))))))))))))......(((((-----...(((((........)))-------------------)).))....)))))))....------ ( -24.40) >DroEre_CAF1 16188 90 - 1 UCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAAGCG-------------------AUCGCUCAGGGCGACAAACG------ ....(((((((((((((((((......)))))))))))))......(((((-----(....))).......(((.-------------------...)))...)))))))....------ ( -25.40) >DroYak_CAF1 10263 90 - 1 UCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAUGCG-------------------AUCGCUCGGGGCGACAAACA------ ....(((((((((((((((((......)))))))))))))......(((.(-----.(.((((((......))))-------------------)).)...).)))))))....------ ( -26.70) >DroAna_CAF1 16651 120 - 1 UCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGCACAGCACAAUUGUACAAUAAAACGAGUGGAUCGCUGAUCACUGAUCGCUGGGGGCGACAGACCGUCGUA ....(((((((((((((((((......)))))))))))))......(((.(.((((.((.((((........)))).))(((((........)))))))))).))))))).......... ( -38.60) >consensus UCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC_____ACAAUUGUACAAUAAAGCG___________________AUCGCCCAGGGCGACAAACG______ ....(((((((((((((((((......)))))))))))))......(((.......((....))........(((.....................)))....))))))).......... (-20.23 = -20.57 + 0.33)
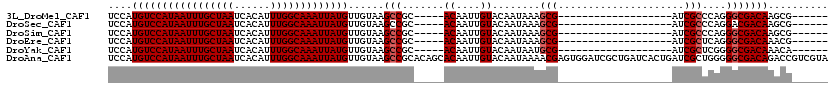
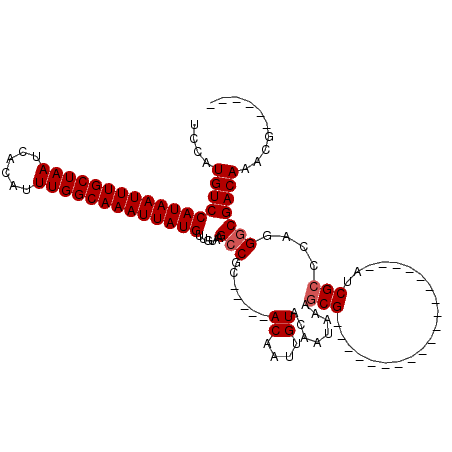
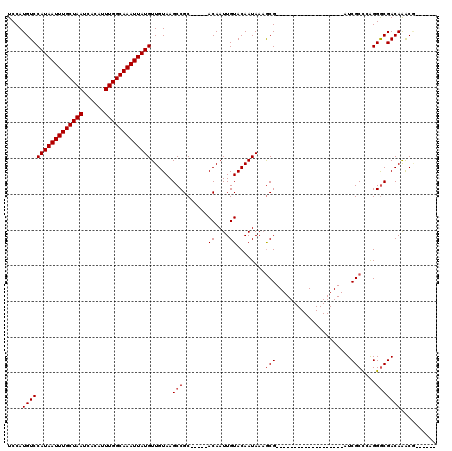
| Location | 1,565,604 – 1,565,706 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -15.99 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1565604 102 - 23771897 UCCAAGCUGCG-------UGCUCCAAGCUACGAGUGC-CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAAGCG----- .....(((.((-------(((.....)).))).((((-.((..(((..(((((((((((((......))))))))))))).)))..)).))-----)).............))).----- ( -28.50) >DroSec_CAF1 10085 90 - 1 UCCAAGCUGCG-------------------CGAGUGC-CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAAGCG----- .....((((..-------------------(((((((-.((..(((..(((((((((((((......))))))))))))).)))..)).))-----))..)))..).....))).----- ( -23.80) >DroSim_CAF1 10368 90 - 1 UCCAAGCUGCG-------------------CGAGUGC-CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAAGCG----- .....((((..-------------------(((((((-.((..(((..(((((((((((((......))))))))))))).)))..)).))-----))..)))..).....))).----- ( -23.80) >DroEre_CAF1 16208 109 - 1 UCCAAGCCCCAAGCCCCAAGCUCCCAGCUGCGAGUGC-CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAAGCG----- .....((.....((....(((.....)))))..((((-.((..(((..(((((((((((((......))))))))))))).)))..)).))-----))..............)).----- ( -26.00) >DroYak_CAF1 10283 102 - 1 UCCAAGCGGCG-------AGCUCCAAGCUGCGAGUGC-UGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC-----ACAAUUGUACAAUAAUGCG----- .....(((((.-------........)))))..((((-.((..(((..(((((((((((((......))))))))))))).)))..)).))-----)).................----- ( -29.50) >DroAna_CAF1 16691 100 - 1 CAUG-GAUGUC-------------------CACGUCCAUGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGCACAGCACAAUUGUACAAUAAAACGAGUGG ((((-((((..-------------------..)))))))).((((...(((((((((((((......)))))))))))))((((..((......))))))................)))) ( -29.10) >consensus UCCAAGCUGCG___________________CGAGUGC_CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUGUUGUAAGCCGC_____ACAAUUGUACAAUAAAGCG_____ .....(((........................................(((((((((((((......)))))))))))))(((((..................)))))...)))...... (-15.99 = -16.10 + 0.11)

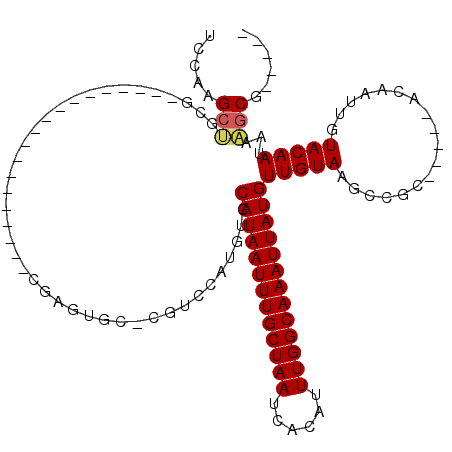
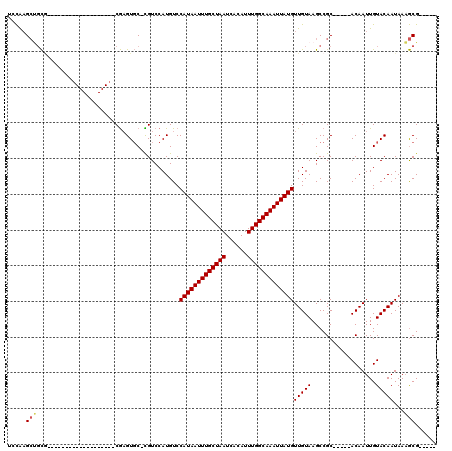
| Location | 1,565,634 – 1,565,746 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.85 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1565634 112 - 23771897 AUUUGCCUCAGUUAAACGAAGCGUUGAAUUGAAAGCGAGCUCCAAGCUGCG-------UGCUCCAAGCUACGAGUGC-CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUG .......((((((.((((...)))).))))))..((.(((.....)))))(-------..(((........)))..)-..........(((((((((((((......))))))))))))) ( -30.70) >DroSec_CAF1 10115 100 - 1 AUUUGCCGCAGUUAAACGAAGCGUUGAAUUGAAAGCGAGCUCCAAGCUGCG-------------------CGAGUGC-CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUG ((((((.((((((........((((........)))).......)))))))-------------------)))))..-..........(((((((((((((......))))))))))))) ( -29.66) >DroSim_CAF1 10398 100 - 1 AUUUGCCGCAGUUAAACGAAGCGUUGAAUUGAAAGCGAGCUCCAAGCUGCG-------------------CGAGUGC-CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUG ((((((.((((((........((((........)))).......)))))))-------------------)))))..-..........(((((((((((((......))))))))))))) ( -29.66) >DroEre_CAF1 16238 119 - 1 AUUUGCCACAGUUAAGCGAAGCGUUGAAUUGAAAGCGAGCUCCAAGCCCCAAGCCCCAAGCUCCCAGCUGCGAGUGC-CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUG ((((((..(((((.((((...)))).)))))..((((((((....((.....))....)))))...)))))))))..-..........(((((((((((((......))))))))))))) ( -31.60) >DroYak_CAF1 10313 112 - 1 AUUUGCCACAGUUAAACGAAGCGUUGAAUUGAAAGCGAGCUCCAAGCGGCG-------AGCUCCAAGCUGCGAGUGC-UGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUG .......(((((....((.((((((........)))((((((........)-------)))))...))).))...))-))).......(((((((((((((......))))))))))))) ( -33.70) >DroAna_CAF1 16731 100 - 1 AUUUACUUCAGUUUAACGAAGCGUUGAAUUGAAAGCGAACCAUG-GAUGUC-------------------CACGUCCAUGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUG ......((((((((((((...)))))))))))).......((((-((((..-------------------..))))))))........(((((((((((((......))))))))))))) ( -35.60) >consensus AUUUGCCACAGUUAAACGAAGCGUUGAAUUGAAAGCGAGCUCCAAGCUGCG___________________CGAGUGC_CGUCCAUGUCCAUAAUUUGCUAAUCACAUUUGGCAAAUUAUG .(((((..(((((.((((...)))).)))))...))))).................................................(((((((((((((......))))))))))))) (-21.33 = -21.22 + -0.11)

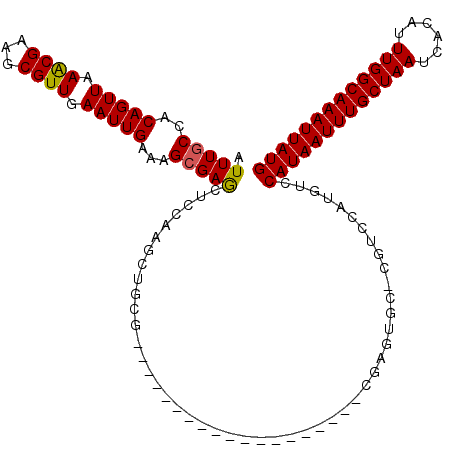
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:42 2006