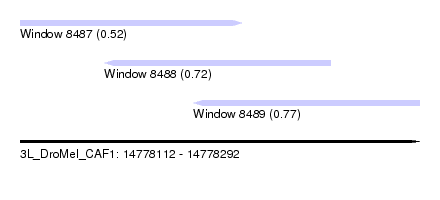
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,778,112 – 14,778,292 |
| Length | 180 |
| Max. P | 0.765169 |

| Location | 14,778,112 – 14,778,212 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14778112 100 + 23771897 AAAGGGGUAU--CAAAUGGGGGAGGGGGGUGGUGUGUAGGGCACUGUGCGGUUGCGGCCUGAAGAUUUUGAAGUUUUUGCGGUGGCCGCUGCUCUUUUCGUU ..........--.........((((((((..(((..(((....)))..)(((..(.((..(((((((....))))))))).)..)))))..))))))))... ( -33.30) >DroSec_CAF1 20974 80 + 1 AAAGGGGUAU--CAAAGGGGGG--------------------ACCGUGCUGUUGCGGCCUGAAGAUUUUGAAGUUUUUGCGGUGGCCGCUGCUCGUUUCGUU (((.(((((.--.....((...--------------------.))........((((((.(((((((....))))))).....))))))))))).))).... ( -23.70) >DroEre_CAF1 20329 82 + 1 AAAGGGGUUUUCCAAGGGGGGG--------------------GCUGUGCGGUUGCGGCCUGAAGAUUUUGAAGUUUUUGCGGUGGCCGCUGCUCGUUUCGUU ...(((....)))..(..(.((--------------------((...(((((..(.((..(((((((....))))))))).)..))))).)))).)..)... ( -31.00) >DroYak_CAF1 21123 82 + 1 AAAGGGGUUUUCCAAAGGGGGG--------------------ACUGUGCGGUUGCGGCCUGAAGAUUUCGAAGUUUUUGCGGUGGCCGCUGCUCGUUUCGUU ...((((((..((....))..)--------------------))...(((((..(.((..(((((((....))))))))).)..)))))..)))........ ( -28.70) >consensus AAAGGGGUAU__CAAAGGGGGG____________________ACUGUGCGGUUGCGGCCUGAAGAUUUUGAAGUUUUUGCGGUGGCCGCUGCUCGUUUCGUU (((.((((.........(.((......................)).)(((((..(.((..(((((((....))))))))).)..))))).)))).))).... (-19.29 = -19.35 + 0.06)

| Location | 14,778,150 – 14,778,252 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -27.73 |
| Consensus MFE | -22.32 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14778150 102 - 23771897 UUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAAAGAGCAGCGG------------------CCACCGCAAAAACUUCAAAAUCUUCAGGCCGCAACCGCACAGUGC ((((((....)))))).....(((.((((((((....((...))..))))...((((------------------((........................))))))...)))).))).. ( -28.06) >DroSec_CAF1 20994 100 - 1 UUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUCAAGCGAAACGAAACGAGCAGCGG------------------CCACCGCAAAAACUUCAAAAUCUUCAGGCCGCAACAGCACGGU-- ((((((....)))))).......(((((........)))))......((.((.((((------------------((........................))))))....)).))..-- ( -25.66) >DroEre_CAF1 20351 100 - 1 CUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAACGAGCAGCGG------------------CCACCGCAAAAACUUCAAAAUCUUCAGGCCGCAACCGCACAGC-- .(((((....))))).........(((((.(((((((((........))....((((------------------...))))..............))))))).))))).........-- ( -26.30) >DroYak_CAF1 21145 100 - 1 CUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAACGAGCAGCGG------------------CCACCGCAAAAACUUCGAAAUCUUCAGGCCGCAACCGCACAGU-- .(((((....))))).........(((((.(((((((.....((((.......((((------------------...))))......))))....))))))).))))).........-- ( -28.12) >DroAna_CAF1 21073 118 - 1 UUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAACGAACAGCGGCGGCCCAGAUGCCGGCGACCACCGCAAAAACUUCAAAAUCUUCAACCCGUAACCGCACCAC-- ((((((....))))))........(((((..((((((((........))....((((.(((((.......)).).)).))))..............))))))..))))).........-- ( -30.50) >consensus UUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAACGAGCAGCGG__________________CCACCGCAAAAACUUCAAAAUCUUCAGGCCGCAACCGCACAGU__ .(((((....))))).........(((((.(((((((......(((.......((((.....................))))......))).....))))))).)))))........... (-22.32 = -22.40 + 0.08)



| Location | 14,778,190 – 14,778,292 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.93 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -28.70 |
| Energy contribution | -28.42 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14778190 102 - 23771897 GAUCCUUUGAGAUGAAGCCAUCGGUUGCAACUCGCGACCGUUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAAAGAGCAGCGG------------------CCACC ..........((((....))))((((((..(((.....((((.(((.((((((((((......)))))).))))..))).))))....)))..))))------------------))... ( -33.30) >DroSec_CAF1 21032 102 - 1 GAUCCUUUGAGAUGAAGCCAUCGGUUGCAACUCGCGGCCGUUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUCAAGCGAAACGAAACGAGCAGCGG------------------CCACC ........(((.((.((((...)))).)).)))..(((((((((((..(((((((((......)))))).)))...))....((...)).)))))))------------------))... ( -34.90) >DroEre_CAF1 20389 102 - 1 GAUCCUUUGAGAUGAAGCCAUCGGUUGCAACUCGCGGCCGCUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAACGAGCAGCGG------------------CCACC ........(((.((.((((...)))).)).)))..(((((((((((.((((((((((......)))))).))))..))....((...)).)))))))------------------))... ( -38.20) >DroYak_CAF1 21183 102 - 1 GAUCCUUUGAGAUGAAGCCAUCGGUUGCAACUCGCGGCCGCUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAACGAGCAGCGG------------------CCACC ........(((.((.((((...)))).)).)))..(((((((((((.((((((((((......)))))).))))..))....((...)).)))))))------------------))... ( -38.20) >DroAna_CAF1 21111 120 - 1 GACCCUUUGAGAUGAAGCCGUUGGCUGCAACUCGCGGCCGUUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAACGAACAGCGGCGGCCCAGAUGCCGGCGACCACC ..(((...........((((((((((((.....)))))((((.(((.((((((((((......)))))).))))..))).)))).......)))))))(((......))))).)...... ( -42.40) >consensus GAUCCUUUGAGAUGAAGCCAUCGGUUGCAACUCGCGGCCGUUGCGCCUAAGCGCAAGAUAAACUUUGCGUCUUGAAGCGAAACGAAACGAGCAGCGG__________________CCACC ..(((((..(((((.......(((((((.....))))))).(((((....)))))............)))))..))).))........................................ (-28.70 = -28.42 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:29 2006