
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,777,061 – 14,777,347 |
| Length | 286 |
| Max. P | 0.997023 |

| Location | 14,777,061 – 14,777,166 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -41.33 |
| Consensus MFE | -32.60 |
| Energy contribution | -33.40 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14777061 105 + 23771897 GGGGCGCGGCCGCGGUGGG---------------AGUGGAAAUGGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCG .((((((((((......))---------------.((((...(((.((((((..((((.((....)).))))........)))))).)))...))))..........))))....)))). ( -40.80) >DroSec_CAF1 19926 105 + 1 GGGGCGUGGUCGCGGUGGG---------------AGUGGAAAUAGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCG .((((((((.(((((..((---------------...((((((((..((..((((((...))))))..)).))))))))...).)..)).))))))).(((........)))...)))). ( -40.70) >DroEre_CAF1 19382 107 + 1 GGGGCGUGGCUGCGGUGGG-------------GGGGUGGAAAUGGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCG .((((((....((((....-------------...((((...(((.((((((..((((.((....)).))))........)))))).)))...))))..........))))..)))))). ( -43.53) >DroYak_CAF1 20069 120 + 1 GGGGCGUGGCCGCGGCGGGAGUGUAGUGGAGUGGAGUGGAAAUGGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCGCAAGUAGAAAUUCGCUAAUGCCCG .((((((..((((.((....))...))))((((((((((...(((.((((((..((((.((....)).))))........)))))).)))...)))).........)))))).)))))). ( -48.00) >DroAna_CAF1 19488 110 + 1 GCGGCGUGGC---GGAAAGAGUUUUGG-----GGGGGGGGAAU--GACUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGUGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCUCG ((((((((((---((..((((......-----((((.((....--..)).))))((((.((....)).))))....))))....)))))).))))...(((........)))...))... ( -33.60) >consensus GGGGCGUGGCCGCGGUGGG_____________GGAGUGGAAAUGGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCG .(((((((((.........................((((...(((.((((((..((((.((....)).))))........)))))).)))...)))).....(.....)))).)))))). (-32.60 = -33.40 + 0.80)


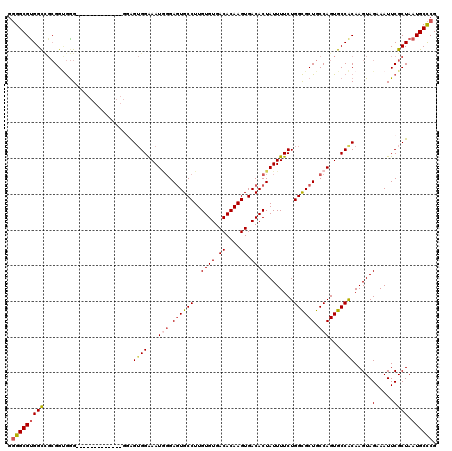
| Location | 14,777,061 – 14,777,166 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -26.02 |
| Energy contribution | -27.10 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14777061 105 - 23771897 CGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCCAUUUCCACU---------------CCCACCGCGGCCGCGCCCC .((((..(((.......)))...(((((.(((((...))))).....(((((..((((((...)))))))))))............---------------.........))))))))). ( -32.00) >DroSec_CAF1 19926 105 - 1 CGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCUAUUUCCACU---------------CCCACCGCGACCACGCCCC .((((....(((...........(((((((((....(....)....)))))))))..(((((......))))).............---------------.....)))......)))). ( -29.60) >DroEre_CAF1 19382 107 - 1 CGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCCAUUUCCACCCC-------------CCCACCGCAGCCACGCCCC .((((..(((.......)))...(((((.(((((...))))).....(((((..((((((...)))))))))))..............-------------.........))))))))). ( -32.60) >DroYak_CAF1 20069 120 - 1 CGGGCAUUAGCGAAUUUCUACUUGCGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCCAUUUCCACUCCACUCCACUACACUCCCGCCGCGGCCACGCCCC .((((..................(((((.(((((...))))).....(((((..((((((...)))))))))))..............................)))))(....))))). ( -34.80) >DroAna_CAF1 19488 110 - 1 CGAGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCACCAGAAAAUAGUGUCACUUGUGUCACACAAGGCAGUC--AUUCCCCCCCC-----CCAAAACUCUUUCC---GCCACGCCGC ((.((..(((.......)))...(((((.((((.....)))).....((((.(.((((((...))))))...).)--)))........-----..............---))))))))). ( -22.70) >consensus CGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCCAUUUCCACUCC_____________CCCACCGCGGCCACGCCCC .((((..(((.......)))...(((((.(((((...))))).....(((((..((((((...)))))))))))....................................))))))))). (-26.02 = -27.10 + 1.08)

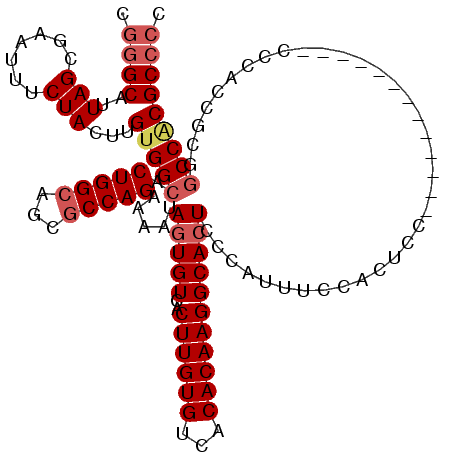
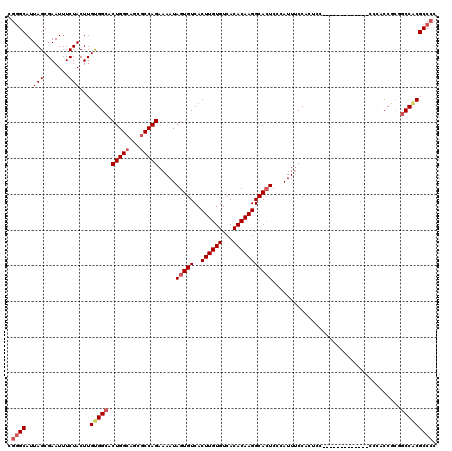
| Location | 14,777,086 – 14,777,197 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.20 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14777086 111 + 23771897 AAUGGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCAC-CAGCA-GCGGCAGCAA------- ......(((((((((((...)))))).).)))).....(((.((((((..(((((...(((........)))...(((.((....)).))))))))-..)))-))).)))...------- ( -44.90) >DroSec_CAF1 19951 111 + 1 AAUAGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCAC-CAGCA-GCGACAGCAG------- ......(((((((((((...)))))).).)))).....(((.((((((..(((((...(((........)))...(((.((....)).))))))))-..)))-))).)))...------- ( -42.40) >DroEre_CAF1 19409 105 + 1 AAUGGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCA----GCAGG----AGCAG------- ..(((((((..((((((...))))))..)).((((((..((((((.....)))))).))))))....)).))).((((((((((((....)))))----)).))----.))).------- ( -38.10) >DroYak_CAF1 20109 113 + 1 AAUGGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCGCAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCAGGCAGCAGGCGGCAGCAA------- ..(((.((((((..((((.((....)).))))........)))))).))).((((((.....(.....).((..((((.(((((.....))))).))))..))))))))....------- ( -46.10) >DroAna_CAF1 19520 116 + 1 AAU--GACUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGUGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCUCGCCGUUGCAGGCGGCAG-UUGCA-ACAACAACAACAUCAAC ..(--((((((.(((((.(.(((..((..(((((.....)))))..))..)))))))))))))......((...(((.((((......))))))).-..)).-............))).. ( -35.50) >consensus AAUGGGAGUGCCUUGUGUGACACAAGUGACACUAUUUUCUGGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCAC_CAGCA_GCGACAGCAA_______ .......(((((.((((...))))(((((..((((((..((((((.....)))))).))))))....)))))...(((.((....)).))))))))........................ (-31.44 = -31.20 + -0.24)


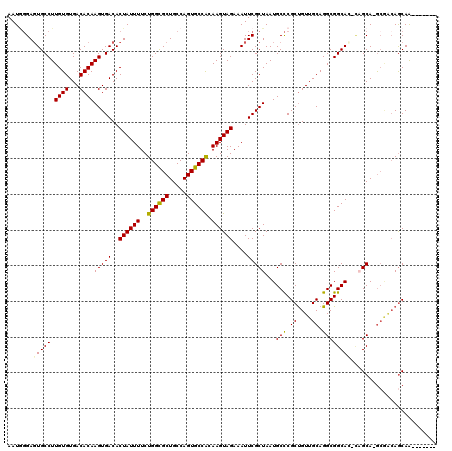
| Location | 14,777,086 – 14,777,197 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -44.64 |
| Consensus MFE | -34.54 |
| Energy contribution | -35.22 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14777086 111 - 23771897 -------UUGCUGCCGC-UGCUG-GUGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCCAUU -------...(((.(((-(((..-(((((((((((....))))))....((((........)))))))))..)))))).))).....(((((..((((((...)))))))))))...... ( -52.00) >DroSec_CAF1 19951 111 - 1 -------CUGCUGUCGC-UGCUG-GUGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCUAUU -------...(((.(((-(((..-(((((((((((....))))))....((((........)))))))))..)))))).))).....(((((..((((((...)))))))))))...... ( -52.00) >DroEre_CAF1 19409 105 - 1 -------CUGCU----CCUGC----UGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCCAUU -------(((..----..(((----((((((((((....))))))....((((........))))......))))))).))).....(((((..((((((...)))))))))))...... ( -35.30) >DroYak_CAF1 20109 113 - 1 -------UUGCUGCCGCCUGCUGCCUGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGCGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCCAUU -------........((((((((((((((((((((....))))))....((((........))))))))..))))))...........((((.((....)).)))).))))......... ( -45.30) >DroAna_CAF1 19520 116 - 1 GUUGAUGUUGUUGUUGU-UGCAA-CUGCCGCCUGCAACGGCGAGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCACCAGAAAAUAGUGUCACUUGUGUCACACAAGGCAGUC--AUU ((((((((..(((((((-((((.-........)))))))))))))))))))((...(((...((((((((.((..((((.........))))..)).))))))))..)))...))--... ( -38.60) >consensus _______UUGCUGCCGC_UGCUG_CUGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCCAGAAAAUAGUGUCACUUGUGUCACACAAGGCACUCCCAUU ..........(((.(((.((((...((((((((((....))))))....((((........))))))))..))))))).))).....(((((..((((((...)))))))))))...... (-34.54 = -35.22 + 0.68)

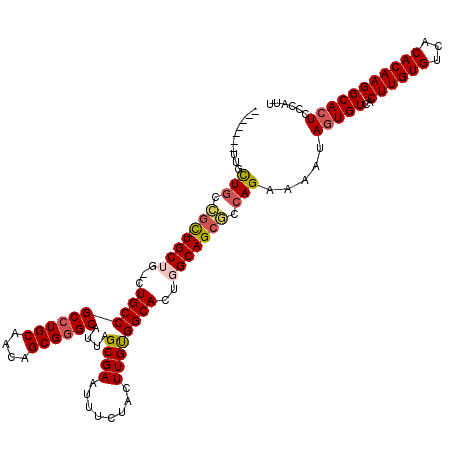
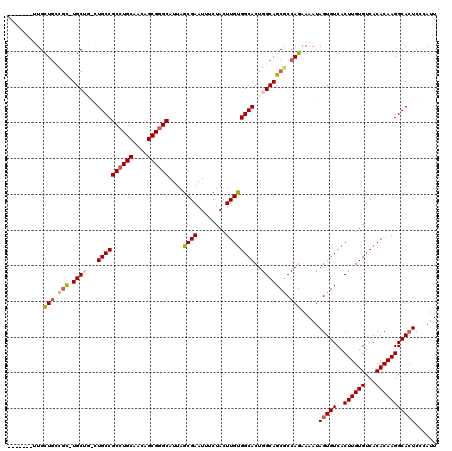
| Location | 14,777,126 – 14,777,230 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.46 |
| Mean single sequence MFE | -43.92 |
| Consensus MFE | -34.44 |
| Energy contribution | -35.80 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14777126 104 + 23771897 GGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCAC-CAGCA-GCGGCAGCAA---------CAACAA----ACAAGCGCACUGG-GGAUGCAGCGCCAU (((((((((((((((...(((........)))..(((..((((((((.((.....)-).)))-))))).))).---------......----....).)))))).-....)))))))).. ( -43.90) >DroSec_CAF1 19991 104 + 1 GGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCAC-CAGCA-GCGACAGCAG---------CAACAA----ACAAGCGCACUGG-GGAUGCAGCGCCAU (((((((((((((((...(((........)))..(((..((((((((..((.....-..)).-)))))))).)---------))....----....).)))))).-....)))))))).. ( -44.70) >DroEre_CAF1 19449 98 + 1 GGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCA----GCAGG----AGCAG---------CAACAA----ACAAGCGCACUGG-GGAUGCAGCGCCAU ((((((((((((((...............(((..((((((((......)))))..----)))..----))).(---------(.....----....)))))))).-....)))))))).. ( -42.90) >DroYak_CAF1 20149 107 + 1 GGCGCUGCCAGUGCCGCAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCAGGCAGCAGGCGGCAGCAA---------CAACAA----ACAAGCGCACUGGGGGAUGCAGCGCCAU ((((((((((((((.((.....(.....)(((..((((.((((((((.....))).))))).))))..)))..---------......----....))))))))......)))))))).. ( -48.90) >DroAna_CAF1 19558 117 + 1 GGUGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCUCGCCGUUGCAGGCGGCAG-UUGCA-ACAACAACAACAUCAACAGCAACGAAAGCGCUAGCACACUGG-GGAUGCCGCGCCAU (((((..((((((.........(.....)((((.((((.(((((.....))))).(-((((.-..................)))))...)))).)))).))))))-((...))))))).. ( -39.21) >consensus GGCGCUGCCAGUGCCACAAGUAGAAAUUCGCUAAUGCCCGCUGUUGCAGGCGGCAC_CAGCA_GCGACAGCAA_________CAACAA____ACAAGCGCACUGG_GGAUGCAGCGCCAU ((((((((((((((...............((....))..((((((((..((........))..))))))))...........................))))))......)))))))).. (-34.44 = -35.80 + 1.36)



| Location | 14,777,126 – 14,777,230 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.46 |
| Mean single sequence MFE | -46.92 |
| Consensus MFE | -36.52 |
| Energy contribution | -36.68 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14777126 104 - 23771897 AUGGCGCUGCAUCC-CCAGUGCGCUUGU----UUGUUG---------UUGCUGCCGC-UGCUG-GUGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCC ..((((((((....-.((((((((((((----((((((---------((((.((.((-.....-..)).))..)))))))))))))..))).........(....))))))))))))))) ( -49.00) >DroSec_CAF1 19991 104 - 1 AUGGCGCUGCAUCC-CCAGUGCGCUUGU----UUGUUG---------CUGCUGUCGC-UGCUG-GUGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCC ...((((((.....-.))))))((..((----.....)---------).))...(((-(((..-(((((((((((....))))))....((((........)))))))))..)))))).. ( -48.20) >DroEre_CAF1 19449 98 - 1 AUGGCGCUGCAUCC-CCAGUGCGCUUGU----UUGUUG---------CUGCU----CCUGC----UGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCC ..((((((((....-.((((((((..((----...(((---------((((.----...))----....((((((....))))))...)))))......))..)).)))))))))))))) ( -43.70) >DroYak_CAF1 20149 107 - 1 AUGGCGCUGCAUCCCCCAGUGCGCUUGU----UUGUUG---------UUGCUGCCGCCUGCUGCCUGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGCGGCACUGGCAGCGCC ..((((((((......((((((((..((----......---------((((((..((((((((..(((.....))).))))))))..))))))......))..)).)))))))))))))) ( -53.00) >DroAna_CAF1 19558 117 - 1 AUGGCGCGGCAUCC-CCAGUGUGCUAGCGCUUUCGUUGCUGUUGAUGUUGUUGUUGU-UGCAA-CUGCCGCCUGCAACGGCGAGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCACC ..((((((((((..-.....))))).)))))...((((((((((((((..(((((((-((((.-........)))))))))))))))))))......(((....)))....))))))... ( -40.70) >consensus AUGGCGCUGCAUCC_CCAGUGCGCUUGU____UUGUUG_________UUGCUGCCGC_UGCUG_CUGCCGCCUGCAACAGCGGGCAUUAGCGAAUUUCUACUUGUGGCACUGGCAGCGCC ..((((((((......((((((((..((....((((((...........((........))........((((((....))))))..))))))......))..)).)))))))))))))) (-36.52 = -36.68 + 0.16)



| Location | 14,777,230 – 14,777,347 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -22.62 |
| Energy contribution | -24.86 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14777230 117 - 23771897 UCGGUG-GGAGGCCUCAUUU--UUUCGCACGCCACGCCUGUGCUUUUGGCGUGUGCUGCUUCCUGCUCAUUGCAUAUCCAACUUUAUUUUUCGCUUCCAGCCGGAAAUAAUAAUUGCAAC (((((.-((((((.......--....(((.((((((((.........)))))).)))))....(((.....)))..................)))))).)))))................ ( -36.50) >DroSec_CAF1 20095 120 - 1 UCGGUGCGGAGGCCUCAUUUUUUUUCGCACGCCACGCCUGUGCUUUUGGCGUGUGCUGCUUCCUGCUCAUUGCAUAUCCAACUUUAUUUUUCGCUUCCAGCCGGAAAUAAUAAUUGCAAC (((((..((((((.............(((.((((((((.........)))))).)))))....(((.....)))..................)))))).)))))................ ( -36.20) >DroEre_CAF1 19547 101 - 1 CCGGAG-UGAGGCCUCAUUU--UUUCGCACGCCACGCCUGUGC----------------UUCCUGCUCAUUGCAUAUCCAACUUUAUUUUUCGCUUCCAGCCGGAAAUAAUAAUUGCAAC ((((..-.(((((.......--....(((((.......)))))----------------....(((.....)))..................)))))...))))................ ( -21.20) >DroYak_CAF1 20256 117 - 1 CCGGAG-UGAGGCCUCAUUU--UUUCGCACGCCACGCCUGUGCUUUUGGCGUGUGCUGCUUCCUGCUCAUUGCAUAUCCAACUUUAUUUUUCGCUUCCAGCCGGAAAUAAUAAUUGCAAC ((((..-.(((((.......--....(((.((((((((.........)))))).)))))....(((.....)))..................)))))...))))................ ( -33.50) >DroAna_CAF1 19675 117 - 1 CCUGUAAGCAGGCCUUAUUU--UUUCACACGCCACGCCUGUGCUU-UGGCGUGUGCUGCUUCCUGCUCAUUGCAUAUCCAACUUUAUUUUUCGCUUCCAGCCGGAAAUAAUAAUUGCAAC ......((((((........--...(((((((((.((....))..-)))))))))......))))))..(((((.........(((((((..((.....))..)))))))....))))). ( -31.75) >consensus CCGGUG_GGAGGCCUCAUUU__UUUCGCACGCCACGCCUGUGCUUUUGGCGUGUGCUGCUUCCUGCUCAUUGCAUAUCCAACUUUAUUUUUCGCUUCCAGCCGGAAAUAAUAAUUGCAAC (((((...(((((............(((((((((.((....))...)))))))))........(((.....)))..................)))))..)))))................ (-22.62 = -24.86 + 2.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:26 2006