
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,769,942 – 14,770,097 |
| Length | 155 |
| Max. P | 0.962603 |

| Location | 14,769,942 – 14,770,059 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.29 |
| Mean single sequence MFE | -22.97 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.99 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14769942 117 - 23771897 CGCAAAGACAAUAAUUCCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAAACCCAAACAAC--AGCAGCAGCAGCGCCAAAA-UAUCUGUGGGGCUUUAACA .((((..((((((......))))))..))))...............((((((.((((....))))....((((......--.((.((....)))).....-......))))))))))... ( -26.67) >DroSec_CAF1 12962 117 - 1 CGCAAAGACAAUAAUUCCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAAACCCAAACAGC--AGCAGCAGCAGCGCCAAAA-UAUCUGUGGGGCUUUAACA .((((..((((((......))))))..))))...............((((((.((((....))))....(((.....((--....)).((((........-...)))))))))))))... ( -27.40) >DroEre_CAF1 12642 120 - 1 CGCAAACACAAUAAUUUCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAGAUCCAAACACCCAAACAGCAGCAGCGCCAAUAAAAUCUGUGGGGCUUUAACA .((((..((((((......))))))..))))...............((((((.((((....))))............((((....((....))..((........))))))))))))... ( -22.30) >DroYak_CAF1 13011 116 - 1 CGCAAACACAAUAAUUCCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAGACCCAAACAGCCAA---GCAGCAGCGCCAAUA-UAUCUGUGGGGCUUUAACA .((((..((((((......))))))..))))...............((((((.((((....))))....(((..((((....---((......)).....-...)))))))))))))... ( -23.10) >DroAna_CAF1 13076 99 - 1 CGCAAAGACAACAAAAGUUUGUUGUUCUUGGCCAACAAAUACAAAAUAAAGCGAA---------CAAAAUCCCAACAAC--AACAU---------GUAUA-CUAUUAUCCAGCUUUAACA .((.((((((((((....)))))).)))).))..............((((((...---------..........(((..--....)---------))...-..........))))))... ( -15.37) >consensus CGCAAAGACAAUAAUUCCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAAACCCAAACAAC__AACAGCAGCAGCGCCAAUA_UAUCUGUGGGGCUUUAACA .((((..((((((......))))))..))))...............((((((.((((....))))....((((............((......))............))))))))))... (-18.24 = -19.23 + 0.99)

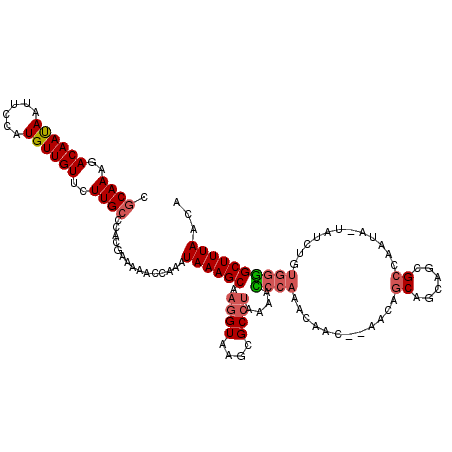
| Location | 14,769,980 – 14,770,097 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.41 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.84 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14769980 117 - 23771897 G--GGGUGGGGGGUUUGCAAAGCUUCGGCAUUGCCGACUGCGCAAAGACAAUAAUUCCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAAACCCAAACAAC- (--(((((((.((((.((((.((....)).)))).)))).)((((..((((((......))))))..))))))))..................((((....))))....))).......- ( -40.20) >DroSec_CAF1 13000 107 - 1 G--GG----------UCCAAAGCUUCGGCAUUGCCGACUGCGCAAAGACAAUAAUUCCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAAACCCAAACAGC- (--((----------(.....((((((((...))))).((.((((..((((((......))))))..)))).))...............))).((((....))))...)))).......- ( -28.40) >DroEre_CAF1 12682 107 - 1 G--GG-----------CCAAAGCUUCGGCAUUGCCGACCGCGCAAACACAAUAAUUUCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAGAUCCAAACACCC (--(.-----------.(......(((((...)))))..((((....((((((......))))))((((((...................))))))...))))....)..))........ ( -24.01) >DroYak_CAF1 13047 109 - 1 GCUAG-----------CUAAAGCUUCGGCAUUGCCGACUGCGCAAACACAAUAAUUCCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAGACCCAAACAGCC (((.(-----------((...((.(((((...)))))..))((((..((((((......))))))..))))..................))).((((....))))...........))). ( -25.00) >DroAna_CAF1 13105 98 - 1 G--AG----------ACCAAAGCUUCGGCAUUGCCGAAUGCGCAAAGACAACAAAAGUUUGUUGUUCUUGGCCAACAAAUACAAAAUAAAGCGAA---------CAAAAUCCCAACAAC- .--..----------......((((((((...)))))..((.(((..(((((((....)))))))..))))).................)))...---------...............- ( -21.00) >consensus G__GG___________CCAAAGCUUCGGCAUUGCCGACUGCGCAAAGACAAUAAUUCCAUGUUGUUCUUGCCCACGAAAAACCAAAUAAAGCAAGGUAAGCGCCUAAAACCCAAACAAC_ .....................((((((((...)))))..(.((((..((((((......))))))..)))).)................))).((((....))))............... (-18.30 = -18.84 + 0.54)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:08 2006