
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,768,585 – 14,768,696 |
| Length | 111 |
| Max. P | 0.944007 |

| Location | 14,768,585 – 14,768,696 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.47 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -16.79 |
| Energy contribution | -16.72 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14768585 111 + 23771897 AUUUGCAAAAUGCAACUAACUGUAAUUAACUGCUUUUUUAAUGCGACCGCAUUAAGAGCGGUUAAAAUUACCGAAAAUU-CUG--------UGAUCAGGCAACUCAUAAAAUCCUAGUCA ...(((.....)))((((.(.((((((((((((((..(((((((....))))))))))))))))..))))).)......-.((--------(((...(....))))))......)))).. ( -26.30) >DroSec_CAF1 11610 111 + 1 AAUUGCGAAAAGUAACUAACUGUAAGUAACUGCGUUUAUAAUGCGACCGCAUUAAGAGCGGUUAAAAUUGCCGAAAAUA-UGA--------AGAUCAGGCAACUCAUAAAAUCCUAUUUA ..((((.....))))....(.((((.(((((((.(((.((((((....))))))))))))))))...)))).).(((((-.((--------..((.((....)).))....)).))))). ( -22.30) >DroSim_CAF1 11767 111 + 1 AAUUGCGAAAAGUAACUAACUGUAAGUAACUGCUUUUAUAAUGCGACCGCAUUAAGAGCGGUUAAAAUUACCGAAAAUG-UAA--------UGAUCAGGCAACUCAUAAAAUCCUAUUUA .((((((...(((.....)))((((.((((((((((((..((((....))))))))))))))))...))))......))-)))--------).((.((....)).))............. ( -24.50) >DroEre_CAF1 11393 111 + 1 AUUUGCGAAAAGCAACUAACUGUAAGUUACUGCUUUGAUAAUGCGACCGCAUUAAGAGCGGUUAAAAUUACCGAAGAUU-CGA--------AGAUCAGGCAACUCAAAAAAUCCUAGUUA ..((((.....)))).(((((((((...((((((((..((((((....)))))))))))))).....))))....((((-...--------.))))((....))...........))))) ( -24.10) >DroYak_CAF1 11590 110 + 1 AUUUGCGAAAAGCAACUAACUGUAAGUAACUGCUUUGAUAAUGCGACCGCAUUAAGAGCGGUUAAAAUUACCGAAGAUU-CGA--------UGAUCAGGCAACUCAUAAAAUCCUAGU-A ...(((.....)))((((.(.((((.((((((((((..((((((....))))))))))))))))...)))).)..((((-..(--------(((...(....)))))..)))).))))-. ( -30.20) >DroAna_CAF1 11155 112 + 1 AAGAGCGAAAAUCCAAUAACUGUAAGAUACAGUUUCGAAAGUGUGACCGCCCCAAAAGCUAUAG-AAUUUUCUAAAAUCGUAAAAGAAGUUUGAUCUGGCAAUAGGUGGAAUC------- ....(((....((....(((((((...)))))))..))...)))..((((((((.(((((.(((-(....))))...((......)))))))....))).....)))))....------- ( -18.20) >consensus AAUUGCGAAAAGCAACUAACUGUAAGUAACUGCUUUGAUAAUGCGACCGCAUUAAGAGCGGUUAAAAUUACCGAAAAUU_CGA________UGAUCAGGCAACUCAUAAAAUCCUAGUUA ..((((.....))))......((((.((((((((((..((((((....))))))))))))))))...)))).........................((....))................ (-16.79 = -16.72 + -0.07)
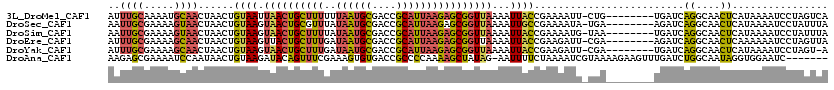

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:05 2006