
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,764,912 – 14,765,036 |
| Length | 124 |
| Max. P | 0.638172 |

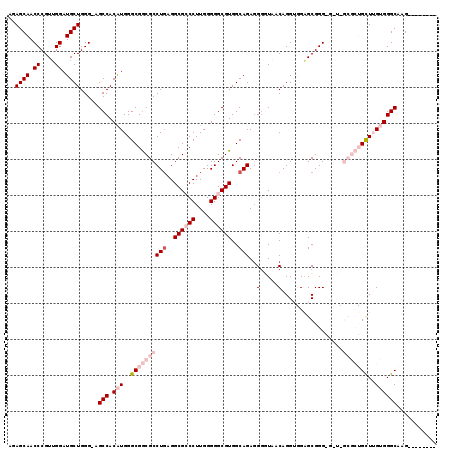
| Location | 14,764,912 – 14,765,022 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.39 |
| Mean single sequence MFE | -46.02 |
| Consensus MFE | -31.98 |
| Energy contribution | -34.74 |
| Covariance contribution | 2.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14764912 110 - 23771897 AGAGCAACCCGUUGGAUGCUGGG-AGCCACAUGGGCGGCGCCUGAGGCGCCCUUGGGGGCGUGGCAGAGGUGUAACAGGUGGAGCGGGGGAUGGCGCUGUUUGUGGGCAAG-------- ..((((.((....)).))))...-.(((.(((..((((((((((..((((((....))))))..))....(((..(....)..)))......))))))))..))))))...-------- ( -47.90) >DroSec_CAF1 7836 110 - 1 AGAGCAACCCGUUGGAUGCUGGGAAGCCACAUGGGCGGCGCCUGAGGCGCCCUUGGGGGCGUGGCAGAGGGGUGACAGGUGGAGCGGGAG-UCGCGCUGUUUGUGGGCAAG-------- ..((((.((....)).)))).....(((.(((..((((((((((..((((((....))))))..)))...(....).....((.(....)-)))))))))..))))))...-------- ( -48.80) >DroSim_CAF1 7972 110 - 1 AGAGCAACCCGUAGGAUGCUGGGAAGCCACAUGGGCGGCGCCUGAGGCGCCCUUGGGGGCGUGGCAGAGGGGUAACAGGUGGAGCGGGAG-UCGCGCUGCUUGUGGGCAAG-------- ..((((.((....)).)))).....(((.((..(((((((((((..((((((....))))))..)))..............((.(....)-))))))))))..)))))...-------- ( -50.40) >DroEre_CAF1 7735 106 - 1 AGAGCAACCCGUUGGAUGCUGGG-AGCCACGUGGGCGGCGCCUGAGGCGGCCUUGGGGGCGUGGCAGAGGGCUAACAGG--GGGCGGG----------GCUGGAGGGCAAGGGUAAAUG ...((..(((((....(.(((..-((((.(....((.((((((((((...))))..)))))).))...)))))..))).--).)))))----------(((....)))....))..... ( -40.90) >DroYak_CAF1 7748 107 - 1 AGAGCAACCCGUUGGAUGCUGGG-AGCCACGUGGGCGACGCCUCUGGCGGCCUUGGGGGCGUGGCAGAUGGUUAACAGGG-GGGCGGG----------GCUGGAGGGCAAGGGUGAAAG ...((..((((((...(.(((..-((((((....((.(((((((..(.....)..))))))).)).).)))))..))).)-.))))))----------(((....)))....))..... ( -42.10) >consensus AGAGCAACCCGUUGGAUGCUGGG_AGCCACAUGGGCGGCGCCUGAGGCGCCCUUGGGGGCGUGGCAGAGGGGUAACAGGUGGAGCGGG_G_U_GCGCUGCUUGUGGGCAAG________ ..((((.((....)).)))).....(((.(((..((((((((((..((((((....))))))..)))..(.....).................)))))))..))))))........... (-31.98 = -34.74 + 2.76)


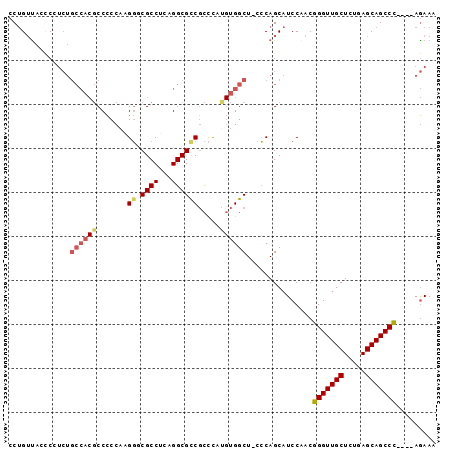
| Location | 14,764,944 – 14,765,036 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 84.46 |
| Mean single sequence MFE | -31.59 |
| Consensus MFE | -23.74 |
| Energy contribution | -24.37 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14764944 92 + 23771897 CCUGUUACACCUCUGCCACGCCCCCAAGGGCGCCUCAGGCGCCGCCCAUGUGGCU-CCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC----AGAAA ...........(((((..(((((....))))).....((.(((((....))))).-))..)).......(((((((.....)))))))----))).. ( -34.40) >DroSec_CAF1 7867 93 + 1 CCUGUCACCCCUCUGCCACGCCCCCAAGGGCGCCUCAGGCGCCGCCCAUGUGGCUUCCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC----AGAAA .(((..........(((.(((((....))))).....)))(((((....)))))....)))........(((((((.....)))))))----..... ( -34.00) >DroSim_CAF1 8003 93 + 1 CCUGUUACCCCUCUGCCACGCCCCCAAGGGCGCCUCAGGCGCCGCCCAUGUGGCUUCCCAGCAUCCUACGGGUUGCUCUGAGCAGCCC----AGAAA ..((((........(((.(((((....))))).....)))(((((....))))).....))))......(((((((.....)))))))----..... ( -34.10) >DroEre_CAF1 7763 92 + 1 CCUGUUAGCCCUCUGCCACGCCCCCAAGGCCGCCUCAGGCGCCGCCCACGUGGCU-CCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC----AGAAA .......((.....((((((.......(((.(((...)))))).....)))))).-....)).......(((((((.....)))))))----..... ( -33.90) >DroYak_CAF1 7777 92 + 1 CCUGUUAACCAUCUGCCACGCCCCCAAGGCCGCCAGAGGCGUCGCCCACGUGGCU-CCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC----AGAAA ...........(((((((((((.....)))((((...))))........))))).-.............(((((((.....)))))))----))).. ( -31.40) >DroPer_CAF1 11574 86 + 1 CCCGAUAGUC---------CCCUCCAAGUACGCCACAGGCGCCAUUUAU-CGGCU-CGCAGCAUCCAAAGGGUUGCUCUGAGCAGCCUUACAAAAAA ..((((((..---------...........((((...))))....))))-))(((-((.((((.((....)).)))).))))).............. ( -21.73) >consensus CCUGUUACCCCUCUGCCACGCCCCCAAGGGCGCCUCAGGCGCCGCCCAUGUGGCU_CCCAGCAUCCAACGGGUUGCUCUGAGCAGCCC____AGAAA ..............((((((.......((.((((...)))))).....))))))...............(((((((.....)))))))......... (-23.74 = -24.37 + 0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:02 2006