
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,762,117 – 14,762,244 |
| Length | 127 |
| Max. P | 0.999909 |

| Location | 14,762,117 – 14,762,216 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -52.32 |
| Consensus MFE | -38.61 |
| Energy contribution | -41.05 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992627 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14762117 99 + 23771897 AUGGCCGC------UGCCGUCGGAGGAAAGUCUGGAAAUGGCCCUGGACCAGGAACCGGC---GGAGGACCCGGUCCUGGUCCAGGCCCACCUCCACACUACGGCGGC ...(((((------((((......))..(((.((((..(((.((((((((((((.((((.---(.....))))))))))))))))).)))..)))).))).))))))) ( -59.50) >DroEre_CAF1 4897 105 + 1 AUGGCCGCUGCAGCUGCCGUUGGAGGAAAGUCUGGAAAUGGCUCUGGACCAGGAACCGGC---GGAGGUCCCGGUCCUGGCCCAGGCCCACCUCCACACUACGGCGGC ...(((((((((((....))))......(((.((((..(((..((((.((((((.((((.---.......)))))))))).))))..)))..)))).))).))))))) ( -52.00) >DroYak_CAF1 4901 105 + 1 AUGGCCGCUGCAGCUGCCGUCGGCGGAAAGUCUGGAAAUGGCCCUGGACCAGGAACCGGC---GGAGGUCCCGGUCCUGGCCCAGGCCCACCUCCACACUACGGCGGC ...(((((((...(((((...)))))..(((.((((..(((.(((((.((((((.((((.---.......)))))))))).))))).)))..)))).))).))))))) ( -57.70) >DroAna_CAF1 4558 99 + 1 AUGGCGGCC---GCCGCAGUGGGCGGAAAGAUCGGCAACGGCCCGGGACCAGGAGCAGGACCGGGAGGACCCG------GCUCAGGACCGCCCCCGCACUUUGGUAGU ..(((....---)))((.(.((((((......((....))((((.......)).)).(..(((((....))))------)..)....))))))).))........... ( -40.10) >consensus AUGGCCGCU___GCUGCCGUCGGAGGAAAGUCUGGAAAUGGCCCUGGACCAGGAACCGGC___GGAGGACCCGGUCCUGGCCCAGGCCCACCUCCACACUACGGCGGC ...(((((((...((((.....))))..(((.((((..(((.(((((.((((((.((((...........)))))))))).))))).)))..)))).))).))))))) (-38.61 = -41.05 + 2.44)

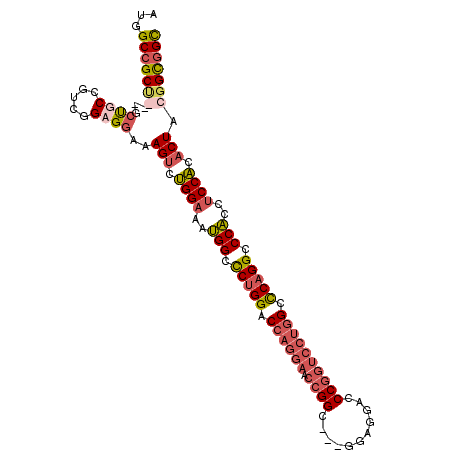
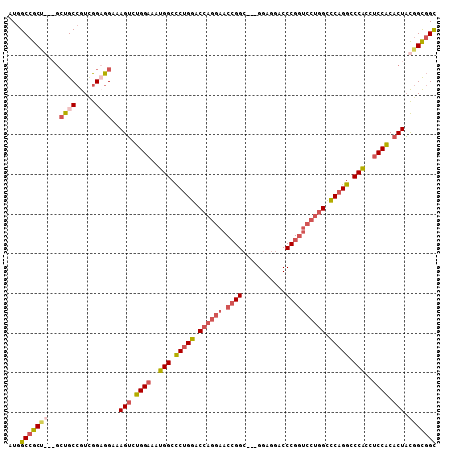
| Location | 14,762,117 – 14,762,216 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.77 |
| Mean single sequence MFE | -59.35 |
| Consensus MFE | -43.66 |
| Energy contribution | -46.98 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.74 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14762117 99 - 23771897 GCCGCCGUAGUGUGGAGGUGGGCCUGGACCAGGACCGGGUCCUCC---GCCGGUUCCUGGUCCAGGGCCAUUUCCAGACUUUCCUCCGACGGCA------GCGGCCAU ((.((((((((.(((((((((.((((((((((((((((.......---.)))).)))))))))))).))))))))).)))........))))).------))...... ( -64.60) >DroEre_CAF1 4897 105 - 1 GCCGCCGUAGUGUGGAGGUGGGCCUGGGCCAGGACCGGGACCUCC---GCCGGUUCCUGGUCCAGAGCCAUUUCCAGACUUUCCUCCAACGGCAGCUGCAGCGGCCAU (((((((((((.(((((((((..(((((((((((((((.......---.)))).)))))))))))..))))))))).)))........))))).((....)))))... ( -61.40) >DroYak_CAF1 4901 105 - 1 GCCGCCGUAGUGUGGAGGUGGGCCUGGGCCAGGACCGGGACCUCC---GCCGGUUCCUGGUCCAGGGCCAUUUCCAGACUUUCCGCCGACGGCAGCUGCAGCGGCCAU (((((.(((((.(((((((((.((((((((((((((((.......---.)))).)))))))))))).)))))))))........(((...))).))))).)))))... ( -69.40) >DroAna_CAF1 4558 99 - 1 ACUACCAAAGUGCGGGGGCGGUCCUGAGC------CGGGUCCUCCCGGUCCUGCUCCUGGUCCCGGGCCGUUGCCGAUCUUUCCGCCCACUGCGGC---GGCCGCCAU ...........((((((((((....((..------(((..(..(((((..((......))..)))))..)...))).))...)))))).))))(((---....))).. ( -42.00) >consensus GCCGCCGUAGUGUGGAGGUGGGCCUGGGCCAGGACCGGGACCUCC___GCCGGUUCCUGGUCCAGGGCCAUUUCCAGACUUUCCGCCGACGGCAGC___AGCGGCCAU (((((...(((.(((((((((.((((((((((((((((...........)))).)))))))))))).))))))))).)))....((.....)).......)))))... (-43.66 = -46.98 + 3.31)



| Location | 14,762,140 – 14,762,244 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.47 |
| Mean single sequence MFE | -52.00 |
| Consensus MFE | -32.35 |
| Energy contribution | -33.35 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14762140 104 + 23771897 GUCUGGAAAUGGCCCUGGACCAGGAACCGGC---GGAGGACCCGGUCCUGGUCCAGGCCCACCUCCACACUACGGCGGCGGC------------GGUGGUGGUGGCGGACACAUGGGAA (((((....(((.((((((((((((.((((.---(.....))))))))))))))))).)))...(((((((((.((....))------------.))))).)))))))))......... ( -59.00) >DroEre_CAF1 4926 101 + 1 GUCUGGAAAUGGCUCUGGACCAGGAACCGGC---GGAGGUCCCGGUCCUGGCCCAGGCCCACCUCCACACUACGGCGGCGGA---------------GGUAGUGGCGGACAUAUGGGUA (((((.....(((.((((.((((((.((((.---.......)))))))))).))))))).((((((.(.(....).)..)))---------------))).....)))))......... ( -48.80) >DroYak_CAF1 4930 101 + 1 GUCUGGAAAUGGCCCUGGACCAGGAACCGGC---GGAGGUCCCGGUCCUGGCCCAGGCCCACCUCCACACUACGGCGGCGGA---------------GGCGGUGGGGGACAUAUGGGCA ((((.....(((.(((((.((((((.((((.---.......)))))))))).))))).)))(((((((.((.((....)).)---------------)...)))))))......)))). ( -49.80) >DroAna_CAF1 4584 113 + 1 GAUCGGCAACGGCCCGGGACCAGGAGCAGGACCGGGAGGACCCG------GCUCAGGACCGCCCCCGCACUUUGGUAGUGGAGGGGGAGGAGGAGGCGGCGGCGGCGGGCAUAUGGGCA ...((....))(((((.(.((....((....((....)).((((------.(((....((.(((((.((((.....))))..))))).))..))).))).)))....)))...))))). ( -50.40) >consensus GUCUGGAAAUGGCCCUGGACCAGGAACCGGC___GGAGGACCCGGUCCUGGCCCAGGCCCACCUCCACACUACGGCGGCGGA_______________GGCGGUGGCGGACAUAUGGGCA (((((((..(((.(((((.((((((.((((...........)))))))))).))))).)))..)))...((((.((......................)).)))).))))......... (-32.35 = -33.35 + 1.00)

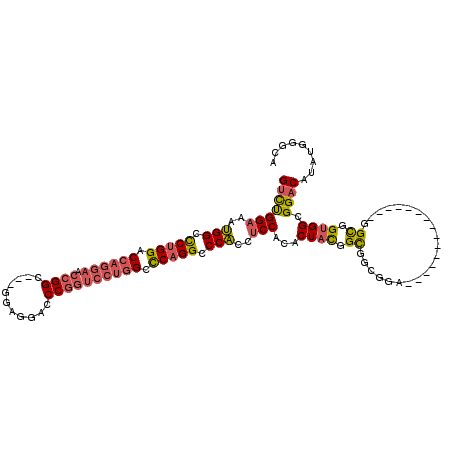
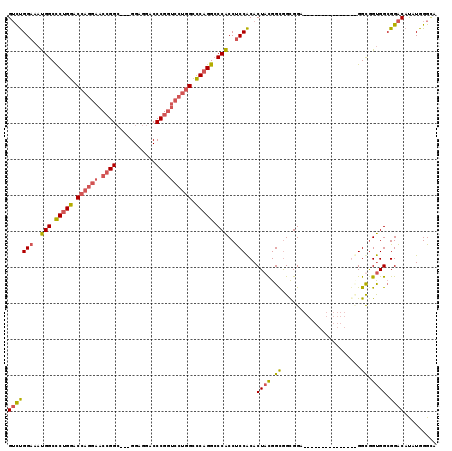
| Location | 14,762,140 – 14,762,244 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.47 |
| Mean single sequence MFE | -51.27 |
| Consensus MFE | -37.81 |
| Energy contribution | -39.88 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 4.23 |
| SVM RNA-class probability | 0.999844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14762140 104 - 23771897 UUCCCAUGUGUCCGCCACCACCACC------------GCCGCCGCCGUAGUGUGGAGGUGGGCCUGGACCAGGACCGGGUCCUCC---GCCGGUUCCUGGUCCAGGGCCAUUUCCAGAC .........(((......(((.((.------------((....)).)).)))(((((((((.((((((((((((((((.......---.)))).)))))))))))).)))))))))))) ( -55.70) >DroEre_CAF1 4926 101 - 1 UACCCAUAUGUCCGCCACUACC---------------UCCGCCGCCGUAGUGUGGAGGUGGGCCUGGGCCAGGACCGGGACCUCC---GCCGGUUCCUGGUCCAGAGCCAUUUCCAGAC .........(((......((((---------------((((((......).)))))))))((((((((((((((((((.......---.)))).))))))))))).))).......))) ( -51.70) >DroYak_CAF1 4930 101 - 1 UGCCCAUAUGUCCCCCACCGCC---------------UCCGCCGCCGUAGUGUGGAGGUGGGCCUGGGCCAGGACCGGGACCUCC---GCCGGUUCCUGGUCCAGGGCCAUUUCCAGAC .((((............(((((---------------((((((......).))))))))))...((((((((((((((.......---.)))).))))))))))))))........... ( -54.60) >DroAna_CAF1 4584 113 - 1 UGCCCAUAUGCCCGCCGCCGCCGCCUCCUCCUCCCCCUCCACUACCAAAGUGCGGGGGCGGUCCUGAGC------CGGGUCCUCCCGGUCCUGCUCCUGGUCCCGGGCCGUUGCCGAUC .........(((((..((((..((.....((.(((((..((((.....)))).))))).))....(..(------((((....)))))..).))...))))..)))))........... ( -43.10) >consensus UGCCCAUAUGUCCGCCACCACC_______________UCCGCCGCCGUAGUGUGGAGGUGGGCCUGGGCCAGGACCGGGACCUCC___GCCGGUUCCUGGUCCAGGGCCAUUUCCAGAC .................................................((.(((((((((.((((((((((((((((...........)))).)))))))))))).))))))))).)) (-37.81 = -39.88 + 2.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:57 2006