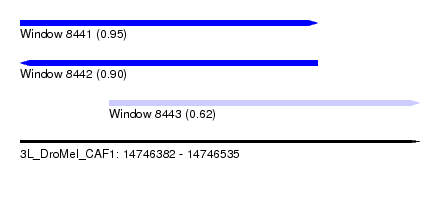
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,746,382 – 14,746,535 |
| Length | 153 |
| Max. P | 0.952713 |

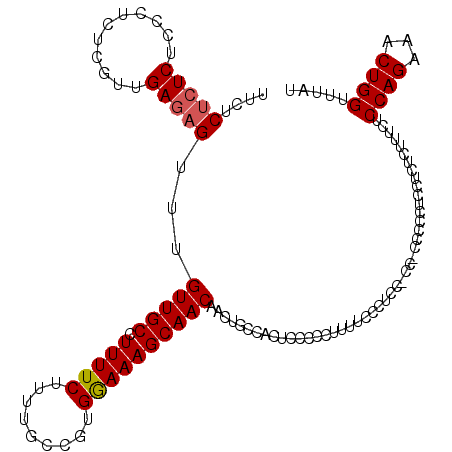
| Location | 14,746,382 – 14,746,496 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952713 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14746382 114 + 23771897 AUAAACCAGUUUCUGGAGAAAGAGAGGAGGGGG------AGGGAAAAGGGGGAGUGGCAGUUGUUGCUUUCCACGGCAAAAGAAAAGGCAACAAACUCUCAACGAGAGGGAGAGAGAGAA .....((..(((((........)))))..))..------.........((..((..((....))..))..))..............(....)...(((((..(....)...))))).... ( -28.40) >DroSec_CAF1 11493 120 + 1 AUAAACCAGUUUCUGGAGAAAGAGAGGAGGGGGAGGAGGAGGAAAAAGGGGGAGUGGCAGUUGUUGCUUUCCACGGUAAAAGAAAAGGCAACAAACUCUCAACGAAGGCGAGAGAGAGAA ....(((..(((((......))))).......................((..((..((....))..))..))..))).........(....)...(((((..(....).)))))...... ( -27.50) >DroEre_CAF1 11781 111 + 1 AUAAACCAGUUUCUGGGC-------CGAGCGGCGGGGGGAGAGAAAAGGGGGAGUGGCAGUUGUUGCUUUUCAUGGCAAAAGAAAAGGCAACGGAC--UCAACGAGAGGGAGAGAGAGAA .....((..((((...((-------(....)))..(..(((.......(..(((..((....))..)))..)..............(....)...)--))..))))).)).......... ( -26.50) >consensus AUAAACCAGUUUCUGGAGAAAGAGAGGAGGGGG_GG_GGAGGGAAAAGGGGGAGUGGCAGUUGUUGCUUUCCACGGCAAAAGAAAAGGCAACAAACUCUCAACGAGAGGGAGAGAGAGAA .....((((...))))................................((((((..((....))..))))))..............(....)...(((((..(....)...))))).... (-21.69 = -22.13 + 0.45)



| Location | 14,746,382 – 14,746,496 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -18.37 |
| Consensus MFE | -14.09 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14746382 114 - 23771897 UUCUCUCUCUCCCUCUCGUUGAGAGUUUGUUGCCUUUUCUUUUGCCGUGGAAAGCAACAACUGCCACUCCCCCUUUUCCCU------CCCCCUCCUCUCUUUCUCCAGAAACUGGUUUAU ....(((((...........))))).(((((((.(((((.........)))))))))))).....................------.................((((...))))..... ( -17.90) >DroSec_CAF1 11493 120 - 1 UUCUCUCUCUCGCCUUCGUUGAGAGUUUGUUGCCUUUUCUUUUACCGUGGAAAGCAACAACUGCCACUCCCCCUUUUUCCUCCUCCUCCCCCUCCUCUCUUUCUCCAGAAACUGGUUUAU ..(((((...((....))..))))).(((((((.(((((.........))))))))))))............................................((((...))))..... ( -18.90) >DroEre_CAF1 11781 111 - 1 UUCUCUCUCUCCCUCUCGUUGA--GUCCGUUGCCUUUUCUUUUGCCAUGAAAAGCAACAACUGCCACUCCCCCUUUUCUCUCCCCCCGCCGCUCG-------GCCCAGAAACUGGUUUAU ............(((.....))--)...(((((.(((((.........)))))))))).............................(((....)-------))((((...))))..... ( -18.30) >consensus UUCUCUCUCUCCCUCUCGUUGAGAGUUUGUUGCCUUUUCUUUUGCCGUGGAAAGCAACAACUGCCACUCCCCCUUUUCCCUCC_CC_CCCCCUCCUCUCUUUCUCCAGAAACUGGUUUAU ....(((((...........)))))...(((((.(((((.........))))))))))..............................................((((...))))..... (-14.09 = -14.53 + 0.45)


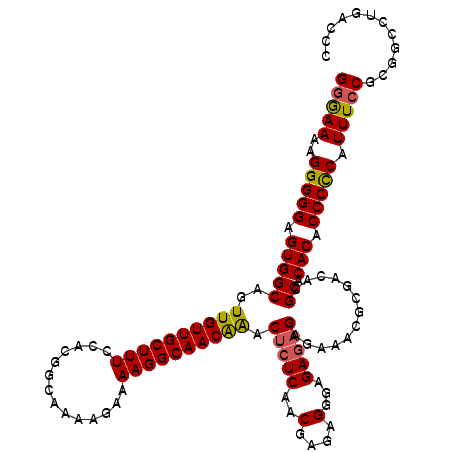
| Location | 14,746,416 – 14,746,535 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -34.45 |
| Energy contribution | -34.57 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14746416 119 + 23771897 GGGAAAAGGGGGAGUGGCAGUUGUUGCUUUCCACGGCAAAAGAAAAGGCAACAAACUCUCAACGAGAGGGAGAGAGAGAAACGCGACACGCACACACCCCCAUUUCCGCGGCCUGACCC .(((((.(((((.(((((.((.(((((((((...............(....)...(((((..(....)...))))).)))).))))))))).))).))))).)))))............ ( -43.60) >DroSec_CAF1 11533 119 + 1 GGAAAAAGGGGGAGUGGCAGUUGUUGCUUUCCACGGUAAAAGAAAAGGCAACAAACUCUCAACGAAGGCGAGAGAGAGAAACGCGACACGCACACACCCCCAUUUCCGCGGCCUGACCC (((((..(((((.(((((.((.((((((((((..............(....)...(((((..(....).))))).).)))).))))))))).))).))))).)))))............ ( -40.70) >DroEre_CAF1 11814 116 + 1 GAGAAAAGGGGGAGUGGCAGUUGUUGCUUUUCAUGGCAAAAGAAAAGGCAACGGAC--UCAACGAGAGGGAGAGAGAGAAACGCG-CACGCACACACCCUCAUUUCCGCGGCCUGACCC (.((((.(((((.(((((..(((((((((((..(......)..))))))))))).(--((..(....)...)))...........-...)).))).))))).)))))............ ( -32.60) >consensus GGGAAAAGGGGGAGUGGCAGUUGUUGCUUUCCACGGCAAAAGAAAAGGCAACAAACUCUCAACGAGAGGGAGAGAGAGAAACGCGACACGCACACACCCCCAUUUCCGCGGCCUGACCC (((((..(((((.(((((..((((((((((..............)))))))))).(((((..(....)...))))).............)).))).))))).)))))............ (-34.45 = -34.57 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:45 2006