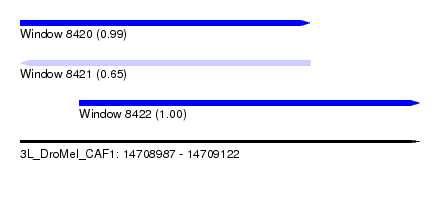
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,708,987 – 14,709,122 |
| Length | 135 |
| Max. P | 0.996607 |

| Location | 14,708,987 – 14,709,085 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.15 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -17.69 |
| Energy contribution | -19.87 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14708987 98 + 23771897 AAGA-------------UUGUACCCAAAUUGGCCAAAGUGCGCAUUUCGU-UCAAGCCCUAUUUUCUGGGCUGGCCAAGUUGCGCU-UUUUGGUCAGCGCAGAAGAAAUGUGG ....-------------...........((((((((((.(((((.((.(.-.((.((((........))))))..).)).))))).-))))))))))((((.......)))). ( -33.80) >DroPse_CAF1 44060 76 + 1 AAGA-------------UUGUAUCCAAAUUGGCCA-AGUGCGCGUUU-----------------------CCGACCAGCUUGCACCGUUUUGGUUAGCUGAGAAGUCGGCUGG ....-------------......(((((.(((.((-(((...((...-----------------------.))....)))))..))).))))).(((((((....))))))). ( -20.40) >DroSim_CAF1 14584 98 + 1 AAGA-------------UUGUACCCAAAUUGGCCACAGUGCGCAUUUCGU-UCAAGCCUUAUUAUCUGGGCUGGCCAAGUUGCGCU-UUUUGGUCAGCGCAGAAGAAAUGUGC ...(-------------((((..((.....))..)))))(((((((((((-....)).......((((.((((((((((.......-.)))))))))).)))).))))))))) ( -38.00) >DroEre_CAF1 14488 109 + 1 CUGAUUUGGUGGCUGAUUUGUACCCGAAUUGGCCGAAGUGCGCGUUUUGU-UCGAGCCUUGUUAUCUG-GCUGGCCAAGUUGCACU--UUUGGCCAGCGCAGAAGAAAUGUGG .........((((..(((((....)))))..)))).....((((((((((-....)).......((((-((((((((((.......--)))))))))).)))).)))))))). ( -40.80) >DroYak_CAF1 15190 99 + 1 AAGA-------------UUGUACCCAAAUUGGCCAGAGUGCGCAUUUCGUUUCGAGCCUUAUUAUCUGGGCUGGCCAAGUUGCAGU-UUUUGGUCAGCGCAGAAGAAAUGUGG ...(-------------((.(..((.....))..).))).((((((((((.....)).......((((.((((((((((.......-.)))))))))).)))).)))))))). ( -32.20) >DroAna_CAF1 15307 84 + 1 AAGA-------------UAGUAUCCAAAUUAGCG-AAGUGCGCACUGGGU-UUGGGCCGUAUAAUUUGGCCUGGCCAAC----------UUGGUCAGCCCAGAAG----GCGC ....-------------.................-....((((.((((((-(.((((((.......))))))((((...----------..)))))))))))...----)))) ( -31.10) >consensus AAGA_____________UUGUACCCAAAUUGGCCAAAGUGCGCAUUUCGU_UCGAGCCUUAUUAUCUGGGCUGGCCAAGUUGCACU_UUUUGGUCAGCGCAGAAGAAAUGUGG ........................................((((((((((.....)).......((((.((((((((((.........)))))))))).)))).)))))))). (-17.69 = -19.87 + 2.17)


| Location | 14,708,987 – 14,709,085 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 69.15 |
| Mean single sequence MFE | -26.47 |
| Consensus MFE | -8.90 |
| Energy contribution | -10.93 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14708987 98 - 23771897 CCACAUUUCUUCUGCGCUGACCAAAA-AGCGCAACUUGGCCAGCCCAGAAAAUAGGGCUUGA-ACGAAAUGCGCACUUUGGCCAAUUUGGGUACAA-------------UCUU (((.........((((((........-))))))..(((((((((((........))))....-.((.....)).....)))))))..)))......-------------.... ( -29.40) >DroPse_CAF1 44060 76 - 1 CCAGCCGACUUCUCAGCUAACCAAAACGGUGCAAGCUGGUCGG-----------------------AAACGCGCACU-UGGCCAAUUUGGAUACAA-------------UCUU ((((((((.....(((((.(((.....)))...)))))((((.-----------------------...)).))..)-)))).....)))......-------------.... ( -20.50) >DroSim_CAF1 14584 98 - 1 GCACAUUUCUUCUGCGCUGACCAAAA-AGCGCAACUUGGCCAGCCCAGAUAAUAAGGCUUGA-ACGAAAUGCGCACUGUGGCCAAUUUGGGUACAA-------------UCUU ((.(((((((((((((((........-))))))....((((..............)))).))-).)))))).))..((((.((.....)).)))).-------------.... ( -30.14) >DroEre_CAF1 14488 109 - 1 CCACAUUUCUUCUGCGCUGGCCAAA--AGUGCAACUUGGCCAGC-CAGAUAACAAGGCUCGA-ACAAAACGCGCACUUCGGCCAAUUCGGGUACAAAUCAGCCACCAAAUCAG ..........((((.(((((((((.--........)))))))))-))))......(((.(((-(............)))))))......(((........))).......... ( -30.80) >DroYak_CAF1 15190 99 - 1 CCACAUUUCUUCUGCGCUGACCAAAA-ACUGCAACUUGGCCAGCCCAGAUAAUAAGGCUCGAAACGAAAUGCGCACUCUGGCCAAUUUGGGUACAA-------------UCUU (((.(((...((((.((((.((((..-........)))).)))).))))......((((.((..((.....))...)).))))))).)))......-------------.... ( -23.30) >DroAna_CAF1 15307 84 - 1 GCGC----CUUCUGGGCUGACCAA----------GUUGGCCAGGCCAAAUUAUACGGCCCAA-ACCCAGUGCGCACUU-CGCUAAUUUGGAUACUA-------------UCUU ((((----((..(((((((.....----------.(((((...)))))......))))))).-....)).))))....-.................-------------.... ( -24.70) >consensus CCACAUUUCUUCUGCGCUGACCAAAA_AGUGCAACUUGGCCAGCCCAGAUAAUAAGGCUCGA_ACGAAAUGCGCACUUUGGCCAAUUUGGGUACAA_____________UCUU ..........((((.((((.((((...........)))).)))).)))).......((((((........((........))....))))))..................... ( -8.90 = -10.93 + 2.03)


| Location | 14,709,007 – 14,709,122 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.79 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -19.71 |
| Energy contribution | -21.60 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.52 |
| SVM decision value | 2.72 |
| SVM RNA-class probability | 0.996607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
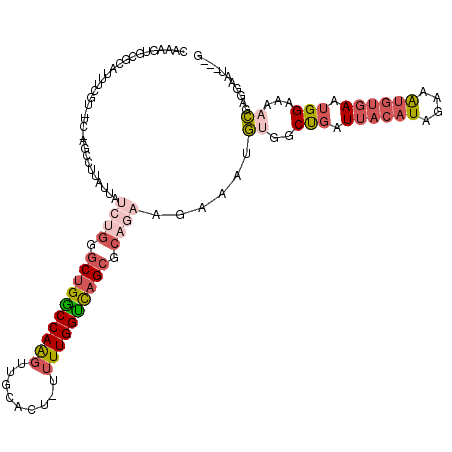
>3L_DroMel_CAF1 14709007 115 + 23771897 CAAAGUGCGCAUUUCGU-UCAAGCCCUAUUUUCUGGGCUGGCCAAGUUGCGCU-UUUUGGUCAGCGCAGAAGAAAUGUGGCUGAUUACAUAGAAAUGUGAAUGGAAAACGAGGAAU---G .......((..((((((-((.(((((..(((((((.((((((((((.......-.)))))))))).)))))))...).))))....((((....))))))))))))..))......---. ( -41.00) >DroSec_CAF1 14908 93 + 1 CACAGUGC--------------------------GGGCUGGCCAAGUUGCGCU-CUUUGGUCAGCGCAGAAGAAAUGUGCCUGAUUACAUAGAAAUGUGAAUGGAAAACGAGGAAUUGUG ((((((.(--------------------------(.((((((((((.......-.)))))))))).)........(((.((...((((((....))))))..))...)))..).)))))) ( -27.90) >DroSim_CAF1 14604 118 + 1 CACAGUGCGCAUUUCGU-UCAAGCCUUAUUAUCUGGGCUGGCCAAGUUGCGCU-UUUUGGUCAGCGCAGAAGAAAUGUGCCUGAUUACAUAGAAAUGUGAAUGGAAAACGAGGAAUGGUG ..(((.(((((((((((-....)).......((((.((((((((((.......-.)))))))))).)))).))))))))))))....((((....))))..................... ( -42.40) >DroEre_CAF1 14521 113 + 1 CGAAGUGCGCGUUUUGU-UCGAGCCUUGUUAUCUG-GCUGGCCAAGUUGCACU--UUUGGCCAGCGCAGAAGAAAUGUGGCCGAUUACAUAGAAAUGUGAAUGGAAAUCGCGGAAU---G .......((((((((..-(((.(((...((.((((-((((((((((.......--)))))))))).)))).)).....))))))...((((....))))....)))).))))....---. ( -42.00) >DroYak_CAF1 15210 116 + 1 CAGAGUGCGCAUUUCGUUUCGAGCCUUAUUAUCUGGGCUGGCCAAGUUGCAGU-UUUUGGUCAGCGCAGAAGAAAUGUGGCCGAUUACAUGGAAAUGUGAAUGGAAAACGGGGAAU---G ...........(..(((((((.(((.((((.((((.((((((((((.......-.)))))))))).))))...)))).))))))((((((....))))))......))))..)...---. ( -41.90) >DroPer_CAF1 44191 91 + 1 CA-AGUGCGCGUUU-----------------------CCGACCAGCUUGCACCGUUUUGGUUAGCUGAGAAGUCGGCUGGCUUGUUA--UGGACGUCCCAAUGGGAAUGGAGGAAA---C ..-.......((((-----------------------((..(((.......((((...((((((((((....))))))))))....)--)))...((((...)))).))).)))))---) ( -31.10) >consensus CAAAGUGCGCAUUUCGU_UC_AGCCUUAUUAUCUGGGCUGGCCAAGUUGCACU_UUUUGGUCAGCGCAGAAGAAAUGUGGCUGAUUACAUAGAAAUGUGAAUGGAAAACGAGGAAU___G ...............................((((.((((((((((.........)))))))))).))))......((..(((.((((((....)))))).)))...))........... (-19.71 = -21.60 + 1.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:25 2006