| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,695,946 – 14,696,046 |
| Length | 100 |
| Max. P | 0.776580 |

| Location | 14,695,946 – 14,696,046 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
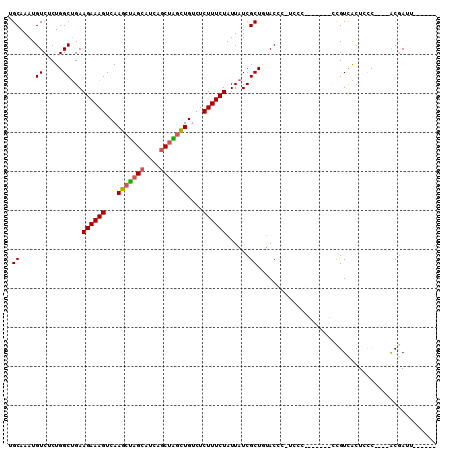
| Mean pairwise identity | 79.04 |
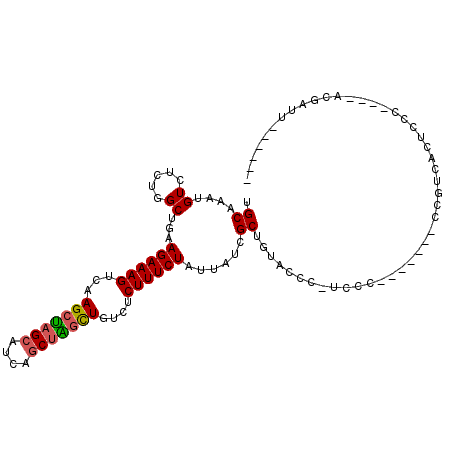
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -16.73 |
| Energy contribution | -16.73 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
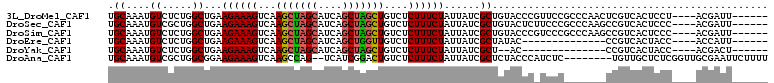
>3L_DroMel_CAF1 14695946 100 - 23771897 UGCAAAUGUCUCUGGCUGAAGAAAGUCAAGCUAGCAUCAGCUAGCUGUCUCUUUCUAUUAUCGCUGUACCCGUUCCGCCCAACUCGUCACUCCU----ACGAUU------ .((.((((..(..(((.((((((((...(((((((....)))))))....))))))....)))))..)..))))..)).....((((.......----))))..------ ( -23.20) >DroSec_CAF1 1808 100 - 1 UGCAAAUGUCGCUGGCUGAAGAAAGUCAAGCUAGCAUCAGCUAGCUGUCUCUUUCUAUUAUCGCUGUACUCUUCCCGCCCAAGCCGUCACUCCC----ACGAUU------ .......((((.(((.(((((((((...(((((((....)))))))....))))))......(((................)))..)))...))----))))).------ ( -25.39) >DroSim_CAF1 1840 100 - 1 UGCAAAUGUCUCUGGCUGAAGAAAGUCAAGCUAGCAUCAGCUAGCUGUCUCUUUCUAUUAUCGCUGUACCCGUCCCGCCCAAGCCGUCACUCCC----ACGAUU------ .............((((..((((((...(((((((....)))))))....))))))......((.(........).))...)))).........----......------ ( -23.50) >DroEre_CAF1 1775 86 - 1 UGCAAAUGUCUCUGGCUGAAGAAAGUCAAGCUAGCAUCAGCUGGUUGUCUCUUUCUAUUAUCGCUAUAC--------------CCGUCACUACC----ACCAUU------ .....(((..(.((((.((((((((...(((((((....)))))))....))))))....)))))).).--------------.))).......----......------ ( -17.70) >DroYak_CAF1 1868 84 - 1 UGCAAAUGUCUCUGGCUGAAGAAAGUCAAGCUAGCAUCAGCUAGCUGUCUCUUUCUAUUAUCGCU--AC--------------CCGUCACUACC----ACGACU------ ............((((.((((((((...(((((((....)))))))....))))))....)))))--).--------------..(((......----..))).------ ( -22.10) >DroAna_CAF1 1456 100 - 1 UGCAAAUGUCGCUGGCGGAAGAAAGUCAAGCCAG--UCAUCGGACUGUCUCUUUCUAUUAUCGCUCUACCCAUCUC--------UGUUGCUCUCGGUUGCGAAUUCUUUU ........((((.(((.((((((((...((.(((--((....))))).))))))))......((...((.......--------.)).))..)).)))))))........ ( -20.70) >consensus UGCAAAUGUCUCUGGCUGAAGAAAGUCAAGCUAGCAUCAGCUAGCUGUCUCUUUCUAUUAUCGCUGUACCC_UCCC_______CCGUCACUCCC____ACGAUU______ .((....((.....))...((((((...(((((((....)))))))....))))))......)).............................................. (-16.73 = -16.73 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:21 2006