
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,688,481 – 14,688,635 |
| Length | 154 |
| Max. P | 0.941711 |

| Location | 14,688,481 – 14,688,595 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.73 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -26.98 |
| Energy contribution | -26.66 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14688481 114 + 23771897 AGUUCGGUAGUCGCAGAAGCAGAA------CAACAGGAACAGCUAGGGGAAAAAUGCUGGAGCUGAGUGGUUGGGAAAUGGGAGAAAAUCUCCAGCUUCGGUUCGAGCAUCAACAUUUUU .((((((.....((.(((((....------((((.....(((((..((........))..)))))....)))).......((((.....)))).))))).))))))))............ ( -32.20) >DroSec_CAF1 37255 113 + 1 AGUUCGGUAGUCCCAGAAGCAGAA------CAACAGGAACAGCUAGGGGAA-AAUGCUGGAGCUGAGUGCCUGGGAAAUGGGGGAAAAUCUCCAGAUUCGGUUCGAGCAUCAACAUUUUU .(((.(((..((((...(((....------...........)))..)))).-...(((.(((((((((.((((.....))))(((.....)))..))))))))).)))))))))...... ( -31.56) >DroSim_CAF1 38405 113 + 1 AGUUCGGUAGUCCCAGAAGCAGAA------CAACAGGAACAGCUAGGGGAA-AAUGCUGGAGCUGAGUGCCUGGGAAAUGGGGGAAAAUCUCCAGCUUCGGUUCGAGCAUCAACAUUUUU .((((((....((..(((((....------...((((..(((((..((...-....))..)))))....)))).....(((((......)))))))))))).))))))............ ( -34.30) >DroEre_CAF1 37207 117 + 1 AGUUCGGCAGUCCCAGAAGCAGAACAGGAACAACAGGAACAGCUAGGGGAA-AAUGCUGGAGCCGAGUGCCUGGGAAAUGG--GAAAAUCUCCAGCUUCGGUUCGAACAUCAACAUUUUU .((((((((.((((...(((.....................)))..)))).-..)))).((((((((.((...(((.((..--....)).))).))))))))))))))............ ( -34.90) >DroYak_CAF1 38088 111 + 1 AGUUCGCUAGUCCCAGAAGCAGAA------CAACAGGAACAGCUAUGGGAA-AGUGCUGGAGCCGAGUGCCUGGGAAAUGG--GAAAAUCUCCAGCUUCGGUUCGAGCAUCAACAUUUUU .(((......(((((..(((....------...........))).))))).-.(((((.((((((((.((...(((.((..--....)).))).)))))))))).))))).)))...... ( -34.76) >consensus AGUUCGGUAGUCCCAGAAGCAGAA______CAACAGGAACAGCUAGGGGAA_AAUGCUGGAGCUGAGUGCCUGGGAAAUGGG_GAAAAUCUCCAGCUUCGGUUCGAGCAUCAACAUUUUU .((((((((.((((...(((.....................)))..))))....)))).((((((((...((((((............)).)))).))))))))))))............ (-26.98 = -26.66 + -0.32)

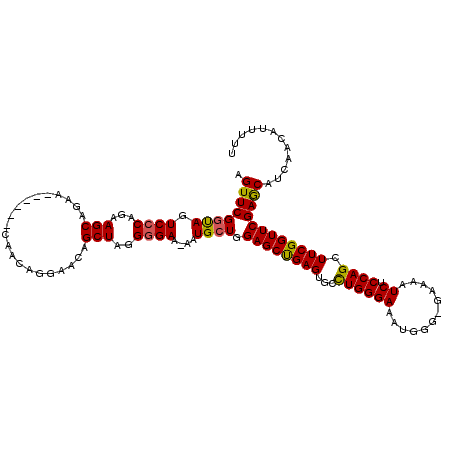

| Location | 14,688,515 – 14,688,635 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.97 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -32.34 |
| Energy contribution | -32.18 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14688515 120 + 23771897 AGCUAGGGGAAAAAUGCUGGAGCUGAGUGGUUGGGAAAUGGGAGAAAAUCUCCAGCUUCGGUUCGAGCAUCAACAUUUUUGGCAGCAAAUAAAAUUUGCUUGUGUUUUCGAGAGAUUUUC .((((((((....(((((.(((((((..((((((((............)).))))))))))))).))))).....))))))))(((((((...))))))).......((....))..... ( -35.10) >DroSec_CAF1 37289 119 + 1 AGCUAGGGGAA-AAUGCUGGAGCUGAGUGCCUGGGAAAUGGGGGAAAAUCUCCAGAUUCGGUUCGAGCAUCAACAUUUUUGGCAGCAAAUAAAAUUUGCUUGUGUUUUCGAGAGAUUUUC .((((((((..-.(((((.(((((((((.((((.....))))(((.....)))..))))))))).))))).....))))))))(((((((...))))))).......((....))..... ( -34.60) >DroSim_CAF1 38439 119 + 1 AGCUAGGGGAA-AAUGCUGGAGCUGAGUGCCUGGGAAAUGGGGGAAAAUCUCCAGCUUCGGUUCGAGCAUCAACAUUUUUGGCAGCAAAUAAAAUUUGCUUGUGUUUUCGAGAGAUUUUC .((((((((..-.(((((.((((((((.((........(((((......))))))))))))))).))))).....))))))))(((((((...))))))).......((....))..... ( -34.30) >DroEre_CAF1 37247 117 + 1 AGCUAGGGGAA-AAUGCUGGAGCCGAGUGCCUGGGAAAUGG--GAAAAUCUCCAGCUUCGGUUCGAACAUCAACAUUUUUGGCAGCAAAUAAAAUUUGCUUGUGUUUUCGAGAGAUUUUC .((((((((..-.(((.(.((((((((.((...(((.((..--....)).))).)))))))))).).))).....))))))))(((((((...))))))).......((....))..... ( -29.80) >DroYak_CAF1 38122 117 + 1 AGCUAUGGGAA-AGUGCUGGAGCCGAGUGCCUGGGAAAUGG--GAAAAUCUCCAGCUUCGGUUCGAGCAUCAACAUUUUUGGCAGCAAAUAAAAUUUGCUUGUGUUUUCGAGAGAUUUUC .(((.(.((((-((((((.((((((((.((...(((.((..--....)).))).)))))))))).))))).....))))).).)))....(((((((.((((......))))))))))). ( -35.10) >consensus AGCUAGGGGAA_AAUGCUGGAGCUGAGUGCCUGGGAAAUGGG_GAAAAUCUCCAGCUUCGGUUCGAGCAUCAACAUUUUUGGCAGCAAAUAAAAUUUGCUUGUGUUUUCGAGAGAUUUUC .((((((((....(((((.((((((((...((((((............)).)))).)))))))).))))).....))))))))(((((((...))))))).......((....))..... (-32.34 = -32.18 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:18 2006