
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,684,960 – 14,685,176 |
| Length | 216 |
| Max. P | 0.992584 |

| Location | 14,684,960 – 14,685,061 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 90.44 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -31.46 |
| Energy contribution | -32.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
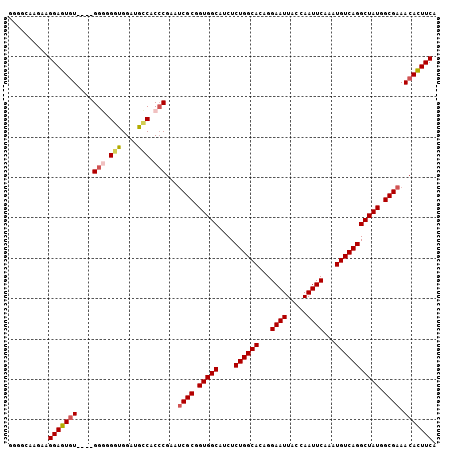
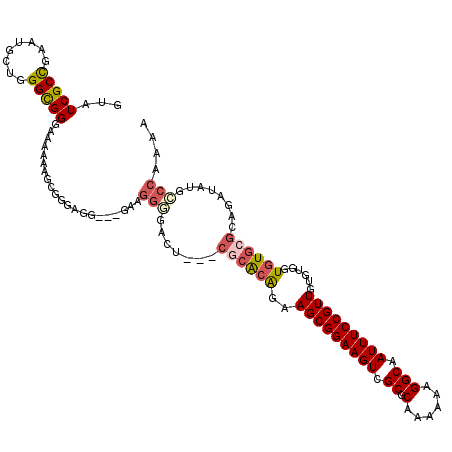
>3L_DroMel_CAF1 14684960 101 - 23771897 GGGGCAAGAAGGAGUAU----GGGGG-CGGAUGACACCCGAAUCGCGGUGGCAUCUCUGGCACAGGAAUUACCAAUUCAAAUGUCAGGCUAUGGCGAAACAUUUCA ...........(((.((----(.(((-.(.....).)))...((((.(((((....((((((...((((.....))))...))))))))))).))))..)))))). ( -30.00) >DroSec_CAF1 35375 102 - 1 GGGGCAAGGAGGAGUGU----GGCUGUUGGUUGGCACCCGAAUCGCGGUGGCAUCUCUGGCACAGGAAUUACCAAUUCAAAUGUCAGGCUAUGGCGAAACACUUCA ..........(((((((----((.((((....)))).))...((((.(((((....((((((...((((.....))))...))))))))))).)))).))))))). ( -35.20) >DroSim_CAF1 36506 102 - 1 GGGGCAAGAAGGAGUGU----GGCUGUUGGUUGGCACCCGAAUCGCGGUGGCAUCUCUGGCACAGGAAUUACCAAUUCAAAUGUCAGGCUAUGGCGAAACACUUCA ..........(((((((----((.((((....)))).))...((((.(((((....((((((...((((.....))))...))))))))))).)))).))))))). ( -35.20) >DroEre_CAF1 35518 99 - 1 GGGGCAU---GGAGUGU----GGGGGGUGGCUGCCACCCGAAACGCGGUGGCAUCUCUGGCACUGGAAUUACCAAUUCAAAUGUCAGGCUAUGGCGAAACACUUCA ......(---(((((((----...((((((...))))))....(((.(((((....((((((...((((.....))))...))))))))))).)))..)))))))) ( -39.90) >DroYak_CAF1 36260 106 - 1 GGGGCAAGAAGGAGUGUUUUGGGGGGGUGGAUGCCACGCGAGUCGCGGUGGCAUCUCUGGCACAGGAAUUACCAAUUCAAAUGUCAGGCUAUGGCGAAACACUUCA ..........(((((((((((....(((((((((((((((...))).)))))))))((((((...((((.....))))...)))))))))....))))))))))). ( -42.90) >consensus GGGGCAAGAAGGAGUGU____GGGGGGUGGAUGCCACCCGAAUCGCGGUGGCAUCUCUGGCACAGGAAUUACCAAUUCAAAUGUCAGGCUAUGGCGAAACACUUCA ..........(((((((....(((.(((....))).)))...((((.(((((....((((((...((((.....))))...))))))))))).)))).))))))). (-31.46 = -32.02 + 0.56)


| Location | 14,685,034 – 14,685,138 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.60 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14685034 104 + 23771897 AUCCG-CCCCCAUACUCCUUC--------UUGCCCCUUUUGGGCAUAUCUACGCACACCACACAACGGAAAUUGCCUUUUUUGCGCGACUUCCGCUUCUGUGCG---AGCCCCCUU ....(-(..............--------.(((((.....)))))......((((((........(((((.(((((......).)))).)))))....))))))---.))...... ( -26.40) >DroSec_CAF1 35449 105 + 1 AACCAACAGCCACACUCCUCC--------UUGCCCCUUUUGGGCAUAUCUACGCACACCAUACAACGGAAAUUGCCUUUUUUGCGCGACUUCCGCUUCUGUGCG---AGUCCCCUU .............((((....--------.(((((.....))))).......(((((........(((((.(((((......).)))).)))))....))))))---)))...... ( -26.60) >DroSim_CAF1 36580 105 + 1 AACCAACAGCCACACUCCUUC--------UUGCCCCUUUUGGGCAUAUCUACGCACACCACACAACGGAAAUUGCCUUUUUUGCGCGACUUCCGCUUCUGUGCG---AGUCCCCUU .............((((....--------.(((((.....))))).......(((((........(((((.(((((......).)))).)))))....))))))---)))...... ( -26.60) >DroEre_CAF1 35592 105 + 1 AGCCACCCCCCACACUCC-----------AUGCCCCUUUCGGCCAUUUCUGCGCACACCACACAACGGAAAUUGCCUUUUUUGCGCGACUUCCGCUUCUGUGCGAACGAACCCCUU ..................-----------..(((......)))...(((..((((((........(((((.(((((......).)))).)))))....))))))...)))...... ( -21.60) >DroAna_CAF1 31721 105 + 1 ----UACCGCCACCAUCCUCCCGAAAAACCAGCCAUAUUUGCGC----CUGCUCAUACCGCACAACGGAAAUUGCCUUUUUUGCGCGACUUCCGCUUUUGUGCG---AGUCUCAGU ----...........................((((....)).))----..(((.(.((((((((((((((.(((((......).)))).)))))...)))))))---.)).).))) ( -23.00) >consensus AACCAACCGCCACACUCCUCC________UUGCCCCUUUUGGGCAUAUCUACGCACACCACACAACGGAAAUUGCCUUUUUUGCGCGACUUCCGCUUCUGUGCG___AGUCCCCUU ..............................(((((.....)))))......((((((........(((((.(((((......).)))).)))))....))))))............ (-18.76 = -19.60 + 0.84)



| Location | 14,685,034 – 14,685,138 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -32.89 |
| Consensus MFE | -22.36 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14685034 104 - 23771897 AAGGGGGCU---CGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGUAGAUAUGCCCAAAAGGGGCAA--------GAAGGAGUAUGGGGG-CGGAU ......(((---(.((((..(((((((((.((.(......))).))))))))).))))..((((.......(((((.....))))).--------......))))..)))-).... ( -34.14) >DroSec_CAF1 35449 105 - 1 AAGGGGACU---CGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUAUGGUGUGCGUAGAUAUGCCCAAAAGGGGCAA--------GGAGGAGUGUGGCUGUUGGUU ....(.(((---((((((..(((((((((.((.(......))).)))))))))......))))).......(((((.....))))).--------....)))).)........... ( -32.50) >DroSim_CAF1 36580 105 - 1 AAGGGGACU---CGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGUAGAUAUGCCCAAAAGGGGCAA--------GAAGGAGUGUGGCUGUUGGUU ....(.(((---((((((..(((((((((.((.(......))).)))))))))......))))).......(((((.....))))).--------....)))).)........... ( -32.50) >DroEre_CAF1 35592 105 - 1 AAGGGGUUCGUUCGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGCAGAAAUGGCCGAAAGGGGCAU-----------GGAGUGUGGGGGGUGGCU ....(((((.(((((((...(((((((((.((.(......))).))))))))).......((((.((....)).((....)).))))-----------...))))))).))..))) ( -33.10) >DroAna_CAF1 31721 105 - 1 ACUGAGACU---CGCACAAAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGCGGUAUGAGCAG----GCGCAAAUAUGGCUGGUUUUUCGGGAGGAUGGUGGCGGUA---- .(((..((.---((((((...((((((((.((.(......))).)))))))))))))))).....)))----.......(((.((((.(..(((....))).).)))).)))---- ( -32.20) >consensus AAGGGGACU___CGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGUAGAUAUGCCCAAAAGGGGCAA________GAAGGAGUGUGGCGGUUGGUU ............((((((...((((((((.((.(......))).)))))))))))))).............(((((.....))))).............................. (-22.36 = -23.20 + 0.84)



| Location | 14,685,061 – 14,685,176 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.28 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -24.34 |
| Energy contribution | -25.01 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14685061 115 - 23771897 GUCUCGCCAAAUGCUGGGUGGGAAAAAAGCGCGAAA--CGAAGGGGGCU---CGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGUAGAUAUGCCCAAAA .(((((((........))))))).............--(....)((((.---((((((..(((((((((.((.(......))).)))))))))......)))))).......)))).... ( -37.20) >DroSec_CAF1 35477 117 - 1 GUAUCGCCGAAUGCUGGGUGGGAAAAAAGCGUGAGGGGAGAAGGGGACU---CGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUAUGGUGUGCGUAGAUAUGCCCAAAA ...((.((..(((((............)))))..)).))...(((.(((---((((((..(((((((((.((.(......))).)))))))))......)))))).))...).))).... ( -37.10) >DroSim_CAF1 36608 117 - 1 GUAUCGCCGAAUGCUGGGUGGGAAAAAAGCGUGAGGGGAGAAGGGGACU---CGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGUAGAUAUGCCCAAAA ...((.((..(((((............)))))..)).))...(((.(((---((((((..(((((((((.((.(......))).)))))))))......)))))).))...).))).... ( -37.10) >DroEre_CAF1 35617 113 - 1 GUAUCGCCGAAUGCUGGGCGGGAAAAAAGCGGA-------AAGGGGUUCGUUCGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGCAGAAAUGGCCGAAA ...((((((....(((.(((.(.....((((((-------......))))))((((((...((((((((.((.(......))).)))))))))))))).).))).)))...))).))).. ( -36.40) >DroYak_CAF1 36366 108 - 1 GUAUCGCCGAAUGCUGGGCGGGAAAAAAGCGGG-------AAGGCGGCUG-----GCGGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGCAGAUAUGGCCGAAA ...((((((.((.(((.(((.(..(..((((((-------((.((((((.-----((....))...))))))((.......))..))))))))...)..).))).))))).))).))).. ( -40.50) >DroAna_CAF1 31753 107 - 1 GUAUCGCU---CGCUGGGCGGGAAAAUUUCGAGCGG---GACUGAGACU---CGCACAAAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGCGGUAUGAGCAG----GCGCAAAU ...(((((---.....)))))...........(((.---..(((..((.---((((((...((((((((.((.(......))).)))))))))))))))).....)))----.))).... ( -35.30) >consensus GUAUCGCCGAAUGCUGGGCGGGAAAAAAGCGGGAGG___GAAGGGGACU___CGCACAGAAGCGGAAGUCGCGCAAAAAAGGCAAUUUCCGUUGUGUGGUGUGCGCAGAUAUGCCCAAAA ...(((((........))))).....................(((.......((((((..(((((((((.((.(......))).)))))))))......))))))........))).... (-24.34 = -25.01 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:16 2006