
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,682,677 – 14,682,944 |
| Length | 267 |
| Max. P | 0.990995 |

| Location | 14,682,677 – 14,682,797 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.46 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14682677 120 - 23771897 UGUGGAAAACGGUUCCGAUUCUGACUCGACGGCUGCAAAUUGUCAGCUCAUGAGUUGCAGCUUAGUUGGCUACAAAAAAAAAAAUGGCCAAAAGAGGCGCAAAAAGAGAACAGAGAAACA ..(((((.....))))).((((((((((..((((((.....).)))))..)))))(((.((((..(((((((............)))))))...))))))).........)))))..... ( -33.60) >DroSec_CAF1 32473 114 - 1 UGUGGAAAACGGUUCCGAUUCUGACUCGACGGCUGCAAAUUGUCCGCUCAUGAGUUGCAGCUCAGUUGGCUA-----AAA-AAAUGGCCAAAAGAGGCGCAAAAAGAGAACAGAGAAACA ..(((((.....))))).(((((.(((((((((........).))).)).....((((..(((..(((((((-----...-...)))))))..)))..))))...)))..)))))..... ( -31.80) >DroSim_CAF1 33633 115 - 1 UGUGGAAAACGGUUCCGAUUCUGACUCGACGGCUGCAAAUUGUCCGCUCAUGAGUUGCAGCUUAGUUGGCUA-----AAAAAAAUGGCCAAAAGAGGCGCAAAAAGAGAACAGAGAAACA ..(((((.....))))).(((((.(((((((((........).))).)).....((((.((((..(((((((-----.......)))))))...))))))))...)))..)))))..... ( -32.20) >DroEre_CAF1 32714 115 - 1 UGUGGAAAACGGUUCCGAUUCUGGCUCGACGGCUGCAAAUUGUCCGCUCAUGAGUUGCAGCUUAGUUGGCUA-----AAAAAAAGGGCCAACAGAGGCGCAAAAAGGGAACAGAGAAACA ...........(((((.....((((.((((((((((((..((......))....))))))))..))))))))-----.......(.(((......))).)......)))))......... ( -33.70) >DroYak_CAF1 33376 115 - 1 UGUGGAAAACGGUUCCGAUUCUGGCUCGACGGCUGCAAAUUGUCCGCUCAUGAGUUGCAGCUUAGUUGGCUA-----AAAAAAAGGGCCAACAGAGGCGCAAAAAGAGAACAGAGAAACA ..(((((.....))))).(((((.(((((((((........).))).)).....((((.((((.(((((((.-----........)))))))..))))))))...)))..)))))..... ( -33.00) >consensus UGUGGAAAACGGUUCCGAUUCUGACUCGACGGCUGCAAAUUGUCCGCUCAUGAGUUGCAGCUUAGUUGGCUA_____AAAAAAAUGGCCAAAAGAGGCGCAAAAAGAGAACAGAGAAACA ..(((((.....))))).(((((.(((....(((((((..((......))....)))))((((..((((((..............))))))...)))))).....)))..)))))..... (-28.42 = -28.46 + 0.04)


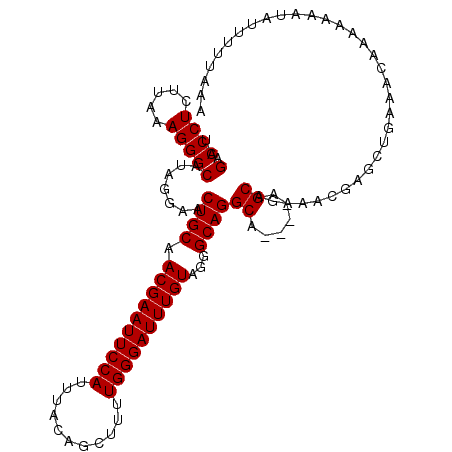
| Location | 14,682,797 – 14,682,904 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14682797 107 - 23771897 UAGACCCUCUUAAAGGGCAUAGGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA----AGCAGAAGCGAGCUGAAACAAAAAAAUAUUUUUAAA ..(.((((.....))))).((((((((((.((((((((((...........))))))))))...))))((.----.((....))..))...............)))))).. ( -26.00) >DroSec_CAF1 32587 107 - 1 UAGACCCUCUUAAAGGGCAUAGUAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA----AGCAGAAACGAGCUGAAACAAAAAAAUAUUUUUAAA ..(.((((.....))))).......((((.((((((((((...........))))))))))...))))...----(((........)))...................... ( -23.40) >DroSim_CAF1 33748 111 - 1 UAGACCCUCUUAAAGGGCAUAGGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCAAGCAAGCAGAAACGAGCUGAAACAAAAAAAUAUUUUUAAA ..(.((((.....))))).((((((((((.((((((((((...........))))))))))...)))).......(((........)))..............)))))).. ( -24.30) >DroEre_CAF1 32829 106 - 1 UAGACCCUCUUAAAGGGCAUAGGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA----AGCAGAAGCGAGCGGAAACAAA-AAAUAUUUUUAAA ..(.((((.....))))).((((((((((.((((((((((...........))))))))))...))))((.----.((....))..))(....)...-.....)))))).. ( -27.90) >DroYak_CAF1 33491 106 - 1 UAGACCCUCUUAAAGGGCAUAUGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA----AGCAGAAAAGAACUGAAACAAA-AAAUAUUUUUAAA ..(.((((.....))))).......((((.((((((((((...........))))))))))...))))...----..(((.......))).......-............. ( -22.20) >consensus UAGACCCUCUUAAAGGGCAUAGGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA____AGCAGAAACGAGCUGAAACAAAAAAAUAUUUUUAAA ..(.((((.....))))).......((((.((((((((((...........))))))))))...))))((......))................................. (-22.70 = -22.70 + -0.00)


| Location | 14,682,833 – 14,682,944 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 95.05 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -25.70 |
| Energy contribution | -25.38 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14682833 111 - 23771897 GAGCGUAAUUUUUUCGUCGUAAUCCAUUUGGAAAAACAAUUAGACCCUCUUAAAGGGCAUAGGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA ..((..................(((..(((......)))...(.((((.....)))))...))).((((.((((((((((...........))))))))))...)))))). ( -27.80) >DroSec_CAF1 32623 111 - 1 GAGCGUAACUUUUUCAUCGCAAUCCCUUUGGAAAAACAAUUAGACCCUCUUAAAGGGCAUAGUAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA ..((.....(((((((............)))))))...((((..((((.....))))..))))..((((.((((((((((...........))))))))))...)))))). ( -27.60) >DroSim_CAF1 33788 111 - 1 GAGCGUAACUUUUUCAUCGCAAUCCCUUUGGAAAAACAAUUAGACCCUCUUAAAGGGCAUAGGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA ..(((............)))..(((..(((......)))...(.((((.....)))))...))).((((.((((((((((...........))))))))))...))))... ( -27.00) >DroEre_CAF1 32864 111 - 1 AAGCGUAACUUUUUCAACAUAAUCCUUUUGGAAAAACAAUUAGACCCUCUUAAAGGGCAUAGGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA ..((..................((((.(((......)))...(.((((.....)))))..)))).((((.((((((((((...........))))))))))...)))))). ( -27.80) >DroYak_CAF1 33526 111 - 1 UAGCGCAACUUUUUCAACGUAAUCCUUUUGGGAAAACAAUUAGACCCUCUUAAAGGGCAUAUGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA ....((...........((((..((((((((((..............))))))))))..))))..((((.((((((((((...........))))))))))...)))))). ( -29.84) >consensus GAGCGUAACUUUUUCAUCGUAAUCCCUUUGGAAAAACAAUUAGACCCUCUUAAAGGGCAUAGGAACUGCAACGAAUUCCAUUUACAGCUUUUGGGAUUUGUAGGGCAGGCA ..((.....(((((((............))))))).......(.((((.....))))).......((((.((((((((((...........))))))))))...)))))). (-25.70 = -25.38 + -0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:13 2006