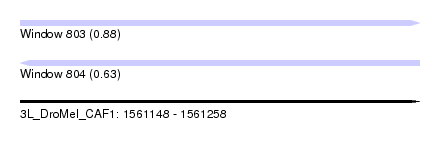
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,561,148 – 1,561,258 |
| Length | 110 |
| Max. P | 0.876705 |

| Location | 1,561,148 – 1,561,258 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -16.13 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
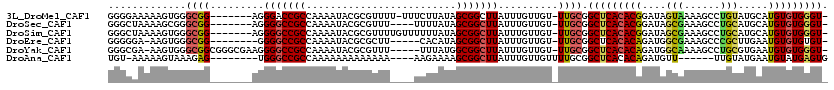
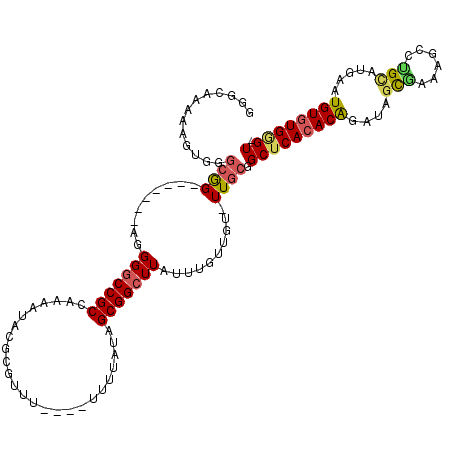
>3L_DroMel_CAF1 1561148 110 + 23771897 -ACCCACACAUGCAUACAGGCUUUUACUAUCCGUGUGAGCCGCAA-ACAACAAAUAAGCCGCUAUAAGAAA-AAAACGCGUAUUUUGGCGGUCCCU-------CCGCCCACUUUUUCCCC -........((((.....(((((.(((.....))).)))))((..-..............)).........-.....)))).....(((((.....-------)))))............ ( -21.89) >DroSec_CAF1 5585 107 + 1 -ACCCACACAUGCAUGCAGGCUUUCGCUAUCCGUGUGAGCCGCAA-ACAACAAAUAAGCCGCUAUAAAA----AAACGCGUAUUUUGGCGGCCCCU-------CCGCCCGCUUUUAGCCC -..........((.(((.(((((.(((.....))).)))))))).-.........(((((((.......----....)))......(((((.....-------))))).))))...)).. ( -27.20) >DroSim_CAF1 5638 111 + 1 -ACCCACACAUGCAUGCAGGCUUUCGCUAUCCGUGUGAGCCGCAA-ACAACAAAUAAGCCGCUAUAAAAAACAAAACGCGUAUUUUGGCGGCCCCU-------CCGCCCACUUUUAGCCC -..........((.(((.(((((.(((.....))).)))))))).-...........(((((((..(((.((.......)).))))))))))....-------..))............. ( -25.70) >DroEre_CAF1 11775 104 + 1 -ACACACACAUUCAAGCGGGCUUUCGCCAUCUGUGUGAGCCGCAA-ACAACAAAUAAGCCGCUAUGUG-----AAGCGCGUAUUUUGGCGGCCCC--------CCGCCCACUU-UCCCCC -............(((.((((..(((((....).))))(((((..-....((((...((((((.....-----.)))).))..)))))))))...--------..)))).)))-...... ( -27.70) >DroYak_CAF1 5752 112 + 1 -ACCCACACAUUCACGCAGGCUUUUGCCAUCUGUGUGAGCCGCAA-ACAACAAAUAAGCCGCCAUAAA-----AAACGCGUAUUUUGGCGGCCCCUUCGCCCGCCGCCCACUU-UCGCCC -..........((((((((((....)))...)))))))((.((..-...........)).))......-----....(((......((((((..........)))))).....-.))).. ( -29.82) >DroAna_CAF1 10681 101 + 1 CACUCAUACAUUCAUACAA------AACAUCUGUGUGAGCCGCAAAACAACAAAUAAGCCGCUUUUCUU----UUUUUUUUUUUUUGGCGGCCCA--------CUCUUUACUUUUU-ACA ..((((((((.........------......))))))))..................((((((......----.............))))))...--------.............-... ( -16.77) >consensus _ACCCACACAUGCAUGCAGGCUUUCGCCAUCCGUGUGAGCCGCAA_ACAACAAAUAAGCCGCUAUAAAA____AAACGCGUAUUUUGGCGGCCCCU_______CCGCCCACUUUUAGCCC ..................(((((.(((.....))).)))))................(((((((.....................)))))))............................ (-16.13 = -16.80 + 0.67)

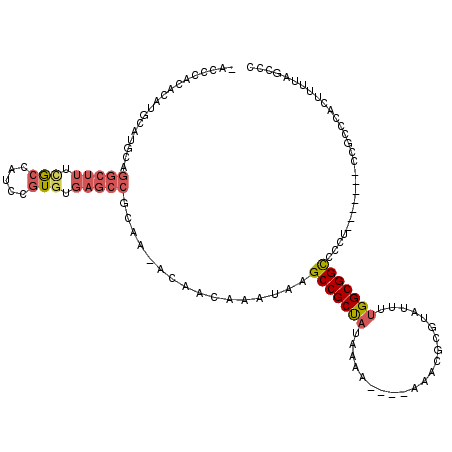
| Location | 1,561,148 – 1,561,258 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.72 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -21.48 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1561148 110 - 23771897 GGGGAAAAAGUGGGCGG-------AGGGACCGCCAAAAUACGCGUUUU-UUUCUUAUAGCGGCUUAUUUGUUGU-UUGCGGCUCACACGGAUAGUAAAAGCCUGUAUGCAUGUGUGGGU- (..(((((.((((((((-------.....)))))......))).))))-)..)....((((((......)))))-)....(((((((((.((((.......)))).....)))))))))- ( -31.30) >DroSec_CAF1 5585 107 - 1 GGGCUAAAAGCGGGCGG-------AGGGGCCGCCAAAAUACGCGUUU----UUUUAUAGCGGCUUAUUUGUUGU-UUGCGGCUCACACGGAUAGCGAAAGCCUGCAUGCAUGUGUGGGU- .......((((.(((((-------.....)))))......(((....----.......))))))).((((((((-(((.(.....).))))))))))).(((..(((....)))..)))- ( -35.70) >DroSim_CAF1 5638 111 - 1 GGGCUAAAAGUGGGCGG-------AGGGGCCGCCAAAAUACGCGUUUUGUUUUUUAUAGCGGCUUAUUUGUUGU-UUGCGGCUCACACGGAUAGCGAAAGCCUGCAUGCAUGUGUGGGU- ..((((...(((.....-------..(((((((((((((....))))))........((((((......)))))-).))))))).)))...))))....(((..(((....)))..)))- ( -34.70) >DroEre_CAF1 11775 104 - 1 GGGGGA-AAGUGGGCGG--------GGGGCCGCCAAAAUACGCGCUU-----CACAUAGCGGCUUAUUUGUUGU-UUGCGGCUCACACAGAUGGCGAAAGCCCGCUUGAAUGUGUGUGU- ......-.....(((((--------....)))))...((((((((((-----((...((((((((..((((..(-(((.(.....).))))..))))))).))))))))).))))))))- ( -40.50) >DroYak_CAF1 5752 112 - 1 GGGCGA-AAGUGGGCGGCGGGCGAAGGGGCCGCCAAAAUACGCGUUU-----UUUAUGGCGGCUUAUUUGUUGU-UUGCGGCUCACACAGAUGGCAAAAGCCUGCGUGAAUGUGUGGGU- ..((..-..((((((.((((((((..(((((((((.((.........-----.)).))))))))).....))))-)))).)))))).(....)))....(((..((......))..)))- ( -43.60) >DroAna_CAF1 10681 101 - 1 UGU-AAAAAGUAAAGAG--------UGGGCCGCCAAAAAAAAAAAAA----AAGAAAAGCGGCUUAUUUGUUGUUUUGCGGCUCACACAGAUGUU------UUGUAUGAAUGUAUGAGUG .((-((((......(((--------((((((((..............----.......)))))))))))....)))))).(((((.(((..(((.------....)))..))).))))). ( -25.40) >consensus GGGCAAAAAGUGGGCGG_______AGGGGCCGCCAAAAUACGCGUUU____UUUUAUAGCGGCUUAUUUGUUGU_UUGCGGCUCACACAGAUAGCGAAAGCCUGCAUGAAUGUGUGGGU_ .............((((.........(((((((.........................)))))))..........)))).(((((((((....(((......))).....))))))))). (-21.48 = -21.54 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:31 2006