
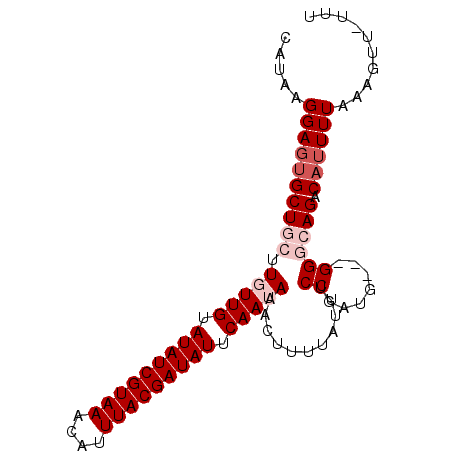
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,682,412 – 14,682,502 |
| Length | 90 |
| Max. P | 0.804080 |

| Location | 14,682,412 – 14,682,502 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -15.74 |
| Energy contribution | -17.34 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
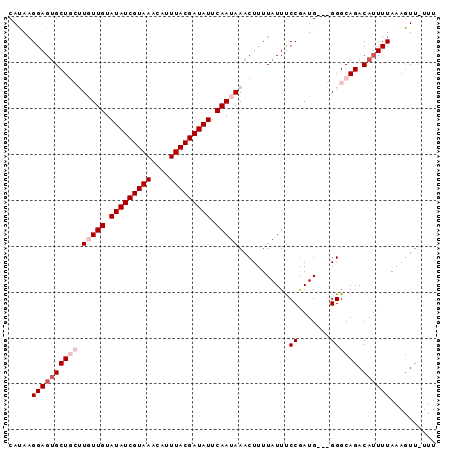
>3L_DroMel_CAF1 14682412 90 + 23771897 CAUAAGGAGUGCUGCUUGUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUG---GGGCAGACAUUUUAAAGUUUUUU .....((((((((((((((((.(((((((((....))))))))).))))))..........((....---)))))).)))))).......... ( -23.90) >DroSec_CAF1 32201 92 + 1 CAUAAGGAGUGCUGCUUGUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUGUGGGGGCAGACAUUUUAAAGUU-UUU .....((((((((((((((((.(((((((((....))))))))).)))))).........(((......))))))).))))))......-... ( -24.70) >DroSim_CAF1 33361 92 + 1 CAUAAGGAGUGCUGCUUGUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUGUGGGGGCAGACAUUUUAAAGUU-UUU .....((((((((((((((((.(((((((((....))))))))).)))))).........(((......))))))).))))))......-... ( -24.70) >DroEre_CAF1 32452 85 + 1 CACA-GGAGGGCU---UCUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUCCCAAUG---GGGCAGACAUUUUAAAGUU-UUU ....-.(((((((---(.(((.(((((((((....))))))))).)))............(((....---)))...........)))))-))) ( -16.00) >DroYak_CAF1 33099 86 + 1 CACAAGGAGUGCU---UCUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUG---GGGCAGACAGUUUAAAGUU-UUU .(((((((....)---))))))(((((((((....)))))))))....((((((.((...(((....---)))..)).)))))).....-... ( -18.10) >consensus CAUAAGGAGUGCUGCUUGUUGUAUAUCGUAAACAUUUACGAUAUUCAAUAAACUUUUAUUUCCGAUG___GGGCAGACAUUUUAAAGUU_UUU .....((((((((((.(((((.(((((((((....))))))))).)))))...........((.......)))))).)))))).......... (-15.74 = -17.34 + 1.60)

| Location | 14,682,412 – 14,682,502 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 92.11 |
| Mean single sequence MFE | -16.24 |
| Consensus MFE | -12.32 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
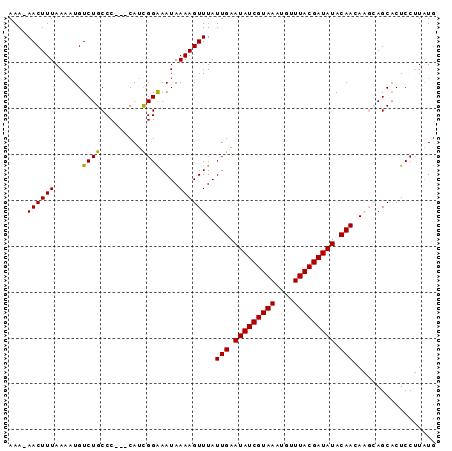
>3L_DroMel_CAF1 14682412 90 - 23771897 AAAAAACUUUAAAAUGUCUGCCC---CAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAACAAGCAGCACUCCUUAUG ..............((.((((((---....)).............(((.(((((((((....))))))))).)))...))))))......... ( -15.60) >DroSec_CAF1 32201 92 - 1 AAA-AACUUUAAAAUGUCUGCCCCCACAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAACAAGCAGCACUCCUUAUG ...-..........((.((((..((.....)).............(((.(((((((((....))))))))).)))...))))))......... ( -16.20) >DroSim_CAF1 33361 92 - 1 AAA-AACUUUAAAAUGUCUGCCCCCACAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAACAAGCAGCACUCCUUAUG ...-..........((.((((..((.....)).............(((.(((((((((....))))))))).)))...))))))......... ( -16.20) >DroEre_CAF1 32452 85 - 1 AAA-AACUUUAAAAUGUCUGCCC---CAUUGGGAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAAGA---AGCCCUCC-UGUG ...-................(((---....))).......((((.(((.(((((((((....))))))))).))).)---))).....-.... ( -15.70) >DroYak_CAF1 33099 86 - 1 AAA-AACUUUAAACUGUCUGCCC---CAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAAGA---AGCACUCCUUGUG ...-..(..((((((.((((...---...))))......))))))..).(((((((((....)))))))))(((((.---.......))))). ( -17.50) >consensus AAA_AACUUUAAAAUGUCUGCCC___CAUCGGAAAUAAAAGUUUAUUGAAUAUCGUAAAUGUUUACGAUAUACAACAAGCAGCACUCCUUAUG ....((((((......((((.........))))....))))))..(((.(((((((((....))))))))).))).................. (-12.32 = -12.00 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:47:10 2006