| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,558,320 – 1,558,412 |
| Length | 92 |
| Max. P | 0.999949 |

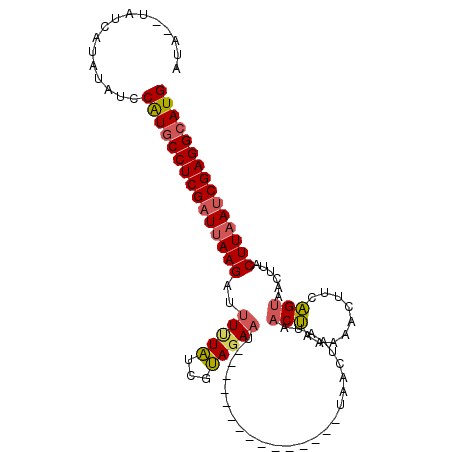
| Location | 1,558,320 – 1,558,412 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -17.98 |
| Energy contribution | -18.35 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.80 |
| SVM decision value | 4.78 |
| SVM RNA-class probability | 0.999949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1558320 92 + 23771897 AUAUAUAUCAUAUAUCCGUGCCUCGAUUAAGAUUUUUAUCGUAGAAU---------------UAACUAAUAACUAAAAUCUUCAGUAACAUACUUAAUCGAGGAAUG ................(((.((((((((((((((((.....(((...---------------...)))......)))))))..((((...))))))))))))).))) ( -17.70) >DroSec_CAF1 2806 90 + 1 AUA--UAUCAUAUAACCAUGCCUCGAUAAAGAUUUGUAUCGUAGAAU---------------UACCUAAUAACUAACAACUUCAGUAACUUACUUAAGCGAGGCAUG ...--...........(((((((((.....(((....))).......---------------.................(((.((((...)))).)))))))))))) ( -18.10) >DroSim_CAF1 2816 90 + 1 AUA--UAUCAUAUAUCCGUGCCUCGAUUAAGAUGUUUAUCGUAGAAU---------------UGCCUAAUAACUAACAACUUCAGUAACUUACUUAAUCGAGGCAUG ...--...........(((((((((((((((.((((.....(((...---------------...)))......))))((....))......))))))))))))))) ( -25.90) >DroEre_CAF1 8883 105 + 1 AUAUAUAUC--ACACCCAUGCCUCGAUUAAGAUUUCUUUCGGAGAAUCACAUAUAGAAUGACUAACUAACAACCAAAUUAUUCGGGAAAUAACUUAAUCGAGGCAUG .........--.....((((((((((((((((((((((..(........).....((((((................)))))))))))))..))))))))))))))) ( -28.29) >consensus AUA__UAUCAUAUAUCCAUGCCUCGAUUAAGAUUUUUAUCGUAGAAU_______________UAACUAAUAACUAAAAACUUCAGUAACUUACUUAAUCGAGGCAUG ................(((((((((((((((..(((((...))))).........................(((.........)))......))))))))))))))) (-17.98 = -18.35 + 0.37)

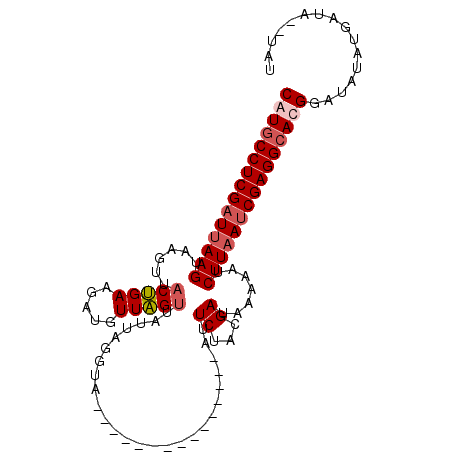
| Location | 1,558,320 – 1,558,412 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.14 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -17.40 |
| Energy contribution | -18.53 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1558320 92 - 23771897 CAUUCCUCGAUUAAGUAUGUUACUGAAGAUUUUAGUUAUUAGUUA---------------AUUCUACGAUAAAAAUCUUAAUCGAGGCACGGAUAUAUGAUAUAUAU ....(((((((((((....(((.((.(((..((((.......)))---------------).))).)).)))....))))))))))).................... ( -18.10) >DroSec_CAF1 2806 90 - 1 CAUGCCUCGCUUAAGUAAGUUACUGAAGUUGUUAGUUAUUAGGUA---------------AUUCUACGAUACAAAUCUUUAUCGAGGCAUGGUUAUAUGAUA--UAU ((((((((......((((((((((..(((........))).))))---------------))).)))((((........))))))))))))...........--... ( -22.30) >DroSim_CAF1 2816 90 - 1 CAUGCCUCGAUUAAGUAAGUUACUGAAGUUGUUAGUUAUUAGGCA---------------AUUCUACGAUAAACAUCUUAAUCGAGGCACGGAUAUAUGAUA--UAU ..(((((((((((((....(((.((.(((((((........))))---------------)))...)).)))....))))))))))))).............--... ( -24.90) >DroEre_CAF1 8883 105 - 1 CAUGCCUCGAUUAAGUUAUUUCCCGAAUAAUUUGGUUGUUAGUUAGUCAUUCUAUAUGUGAUUCUCCGAAAGAAAUCUUAAUCGAGGCAUGGGUGU--GAUAUAUAU (((((((((((((((..(((((((((.....))))...(..(..((((((.......))))))..)..)..)))))))))))))))))))).....--......... ( -32.30) >consensus CAUGCCUCGAUUAAGUAAGUUACUGAAGAUGUUAGUUAUUAGGUA_______________AUUCUACGAUAAAAAUCUUAAUCGAGGCACGGAUAUAUGAUA__UAU (((((((((((((((......(((((.....)))))..........................((...)).......)))))))))))))))................ (-17.40 = -18.53 + 1.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:29 2006