
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,669,521 – 14,669,813 |
| Length | 292 |
| Max. P | 0.931605 |

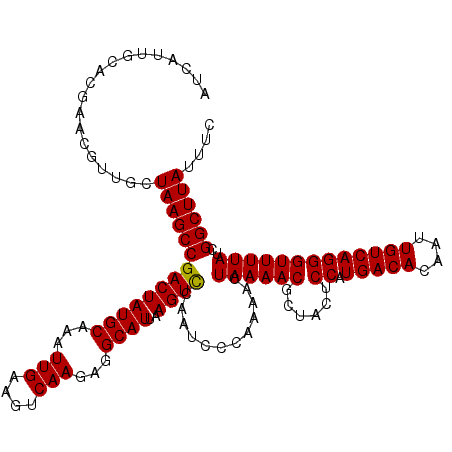
| Location | 14,669,521 – 14,669,641 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.72 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -24.95 |
| Energy contribution | -24.82 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931605 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14669521 120 - 23771897 AUCAUUGCACGAACGUUGCUAAGCCGACUAUGCAAAUUGAAGUCAAGAGGCAUUAAGUCCAAUCCCAAAACUAAAACGCUACUCCAUGACACAAUUGUCAGGGUUUUAUCGGCUUAUUUC ......((((....).)))((((((((((((((...(((....)))...))))..))))............((((((......((.(((((....)))))))))))))..)))))).... ( -26.70) >DroSec_CAF1 19287 120 - 1 AUCAUUGCACGAACGUUGCUAAGCCGACUAUGCAAAUUGAAGUCAAGAGGCAUUAAGUCCAAUCCCAAAACUAAAACGCUACUCCAUGACACAAUUGUCAGGGUUUUAUCGGCUUAUUUC ......((((....).)))((((((((((((((...(((....)))...))))..))))............((((((......((.(((((....)))))))))))))..)))))).... ( -26.70) >DroSim_CAF1 20141 120 - 1 AUCAUUGCACGAACGUUGCUAAGCCGACUAUGCAAAUUGAAGUCAAGAGGCAUUAAGUCCAAUCCCAAAACUAAAACGCUACUCCAUGACACAAUUGUCAGGGUUUUAUCGGCUUAUUUC ......((((....).)))((((((((((((((...(((....)))...))))..))))............((((((......((.(((((....)))))))))))))..)))))).... ( -26.70) >DroEre_CAF1 19772 120 - 1 AUCAUUGCACGAACGUUGCUAAGCCGACUAUGCAAAUUGAAGUCAAGAGGCAUUAAGUCCGAUCACAAAACUAAAACGCUACUCCAUGACACAAUUGUCAGGGUUUUAUCGGCUUAUUUC ......((((....).)))((((((((((((((...(((....)))...))))..))))............((((((......((.(((((....)))))))))))))..)))))).... ( -26.70) >DroYak_CAF1 20019 120 - 1 AUCAUUGCACGAACGUUGCUAAGCCGACUAUGCAAAUUGAAGUCAAGAGGCAUUAAGUCCGAUCCCAAAACUAAAACGCUACUCCAUGACACAAUUGUCAGGGUUUUAUCGGCUUAUUUC ......((((....).)))((((((((((((((...(((....)))...))))..))))............((((((......((.(((((....)))))))))))))..)))))).... ( -26.70) >DroAna_CAF1 16892 120 - 1 AUCAUUCAACUGACGCUGCUAAGCCGACUAUGCAAAUUGAAGUCAAGAGGCAUUAAGUUCGAUCCCCACACUAAAACGCUACUCCAUGACACAAUUGUCAGGGUUUUAUCGGCUUAUUCC ...................((((((((..((((...(((....)))...)))).......((..(((..........(......).(((((....))))))))..)).)))))))).... ( -24.30) >consensus AUCAUUGCACGAACGUUGCUAAGCCGACUAUGCAAAUUGAAGUCAAGAGGCAUUAAGUCCAAUCCCAAAACUAAAACGCUACUCCAUGACACAAUUGUCAGGGUUUUAUCGGCUUAUUUC ...................((((((((((((((...(((....)))...))))..))))............((((((......((.(((((....)))))))))))))..)))))).... (-24.95 = -24.82 + -0.14)


| Location | 14,669,641 – 14,669,737 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 90.36 |
| Mean single sequence MFE | -16.86 |
| Consensus MFE | -14.18 |
| Energy contribution | -13.66 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14669641 96 - 23771897 UCAUCGGUUUAACUUUUCUUCCGUUUCCUCCGAC----CGGAAAAAGCUCUUCCUUAGCUGUCAACUGGUAAAUGCCGAAUUCUAUCAAUUUGUCAUUCA ....(((...(((.........)))....)))((----(((....((((.......)))).....))))).((((.((((((.....)))))).)))).. ( -15.50) >DroSec_CAF1 19407 96 - 1 UCAUCGGUUUAACUUUUCUUCCGUUUCCUCCGAC----CGGAAAAAGCUUUUCCUUAGCUGUCAACUGCUAAAUGCCGAAUUCUAUCAAUUCGUCAUUCA ...((((...(((.........)))....)))).----.(((((.....))))).((((........))))((((.((((((.....)))))).)))).. ( -17.70) >DroSim_CAF1 20261 96 - 1 UCAUCGGUUUAACUUUUCUUCCGUUUCCUCCGAC----CGGAAAAUGCUUUUCCUUAGCUGUCAACUGCUAAAUGCCGAAUUCUAUCAAUUCGUCAUUCA ...((((...(((.........)))....)))).----.(((((.....))))).((((........))))((((.((((((.....)))))).)))).. ( -17.70) >DroEre_CAF1 19892 95 - 1 UCAUCAGUUUAACUUUCCCUCCGUUUCCUCCGUC----CGGAA-AAGCUUUUCCUUAGCUGUCAACUGGUAAGUGCCGAAUUCUAUCAAUUUGUCAUUCA ..(((((((..............(((((......----.))))-)((((.......))))...))))))).((((.((((((.....)))))).)))).. ( -16.20) >DroYak_CAF1 20139 97 - 1 GCAUCAGUUUAA--UUCCUUCCGCUUUCUCCGUCUCACCGGAA-AAGCUUUUCCCUAGCUGUCAACUGCUAAGUGCCGAAUUCUAUCAAUUUGUCAUUCA ((((........--........(((((.((((......)))))-)))).......((((........)))).))))((((((.....))))))....... ( -17.20) >consensus UCAUCGGUUUAACUUUUCUUCCGUUUCCUCCGAC____CGGAAAAAGCUUUUCCUUAGCUGUCAACUGCUAAAUGCCGAAUUCUAUCAAUUUGUCAUUCA ....(((((...................((((......))))...((((.......))))...)))))...((((.((((((.....)))))).)))).. (-14.18 = -13.66 + -0.52)



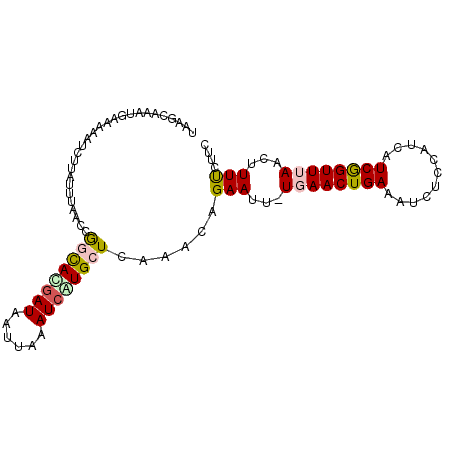
| Location | 14,669,716 – 14,669,813 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -14.70 |
| Consensus MFE | -9.18 |
| Energy contribution | -9.46 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14669716 97 - 23771897 UAGGCAAAUGAAAAGUCCUUUUUAACCGGCACGAUAAUUAAAUCAUGCUCAAACAGAAUU-UGAACUGAAAUCUCCAUCAUCGGUUUAACUUUUCUUC .........(((((((...........((((.(((......))).))))...........-((((((((...........)))))))))))))))... ( -16.40) >DroSec_CAF1 19482 97 - 1 UAAGCAAAUGAAAAAUUUUAUUUAACCGGUAUGAUAAAUAAAUCAUGCUCAAACAGAAUU-UGAACUGAAAUCUGCAUCAUCGGUUUAACUUUUCUUC (((((..((((................((((((((......))))))))....((((.((-(.....))).))))..))))..))))).......... ( -17.00) >DroSim_CAF1 20336 97 - 1 UAAGCAAAUGAAAAAUCUUAUUUAACCGGUACGAUAAUUAAAUCAUGCUCAAACAGAAUU-UGAACUGAAAUCUCCAUCAUCGGUUUAACUUUUCUUC ..(((((((((......)))))).........(((......)))..))).....((((.(-((((((((...........)))))))))...)))).. ( -11.70) >DroEre_CAF1 19966 97 - 1 UAAACAAAUGAAAUAUCAUAUUAAACCAUCAUGAUAAUUAAAUCGUGCUCAAACAGAAUU-UCAACUGAAAUCGCCAUCAUCAGUUUAACUUUCCCUC (((((..((((..................((((((......))))))........((.((-((....))))))....))))..))))).......... ( -11.60) >DroYak_CAF1 20217 84 - 1 UAAACAAA------------UUUAACCAUCACGAUAAUUAAAUGGUGCUCAAAGAGAAUUCUCAACUGAAAUCUCCAGCAUCAGUUUAA--UUCCUUC ........------------...............((((((((((((((....((((.(((......))).)))).))))))).)))))--))..... ( -16.80) >consensus UAAGCAAAUGAAAAAUCUUAUUUAACCGGCACGAUAAUUAAAUCAUGCUCAAACAGAAUU_UGAACUGAAAUCUCCAUCAUCGGUUUAACUUUUCUUC ...........................((((((((......))))))))......(((...((((((((...........))))))))...))).... ( -9.18 = -9.46 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:57 2006