
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,667,548 – 14,667,747 |
| Length | 199 |
| Max. P | 0.993937 |

| Location | 14,667,548 – 14,667,649 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -25.29 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.23 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14667548 101 + 23771897 CCACUUGAAAUGUAUACCCAACAGGGAGUCGAA--ACUGGGACUCCCGACCAUUCCAU---UCAG-------CAACCAGGAUCCAUGAC---AACCCCUCGAGCACCCAU----CUUACA ...(((((...............(((((((...--.....)))))))...((((((..---....-------......)))...)))..---......))))).......----...... ( -18.40) >DroSec_CAF1 17303 101 + 1 CCACUUGAAAUGUAUACCCAACAGGGAGUCGAA--ACUGGGACUCCCGACCAUGCCAU---CCAG-------CAACCAGGAUCCAUGAC---AACCCCUAGAGCACCCAG----CUUCCA ..........(((..........(((((((...--.....)))))))...((((..((---((..-------......)))).))))))---).......((((.....)----)))... ( -23.20) >DroSim_CAF1 18127 101 + 1 CCACUUGAAAUGUAUACCCAACAGGGAGUCGAA--ACUGGGACUCCCGACCAUGCCAU---CCAG-------CAACCAGGAUCCAUGAC---AACCCCUCGAGCACCCAG----CUUCCA ...(((((...............(((((((...--.....)))))))...((((..((---((..-------......)))).))))..---......))))).......----...... ( -23.60) >DroEre_CAF1 17812 111 + 1 CCACUUGAAAUGUAUACCCAACAGGGAGUCGCA--ACUGGGACUCCCGACCAUGCCAUCAUCCAGGCAUGGCAAACCAGGAUCCAUGAC---AACCCCUCGAGCUCCCAG----CUUCCA ....(((..(((.((.((.....(((((((.(.--...).)))))))(.(((((((........))))))))......)))).)))..)---))......((((.....)----)))... ( -34.60) >DroYak_CAF1 18094 101 + 1 CCACUUGAAAUGUAUACCCAACAGGGAGUCGGA--GCUGCGACUCCCAACUAUGCCAU---CCAG-------CAACCAGGAUCCAUGAC---AACCCCUCGAGCACCCAU----CUUCCA ...(((((...............((((((((..--....))))))))...((((..((---((..-------......)))).))))..---......))))).......----...... ( -23.90) >DroAna_CAF1 14816 110 + 1 CCACUUGAAAUGGAUACCCAACAGGGAGUCCCAGUAUUGUGACUCCCAACUGCCCCAU---CCAA-------CCACCAAAUUCUGAAACUGGAGCUCCUGGGUCUCCUGAACUCCUGACU ...........(((.(((((...(((((((.(......).)))))))....((.(((.---....-------.................))).))...))))).)))............. ( -28.05) >consensus CCACUUGAAAUGUAUACCCAACAGGGAGUCGAA__ACUGGGACUCCCGACCAUGCCAU___CCAG_______CAACCAGGAUCCAUGAC___AACCCCUCGAGCACCCAG____CUUCCA ...((((...(((.......)))(((((((..........))))))).............................))))........................................ (-14.04 = -14.23 + 0.20)

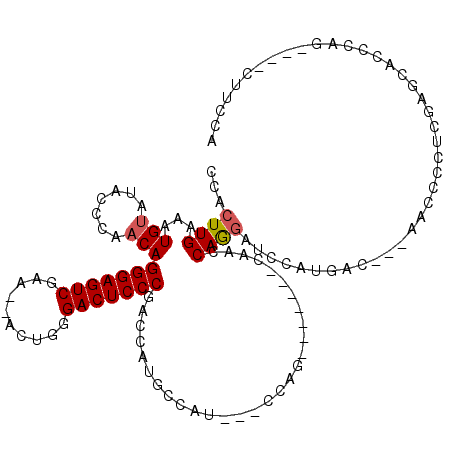

| Location | 14,667,616 – 14,667,714 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.73 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -12.61 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14667616 98 + 23771897 AUCCAUGAC---AACCCCUCGAGCACCCAU----CUUACAGUU--------------CCCUAUUUUUUCGUCCCCAUGGCCGGACUGGAAAACCUGA-AAUGGAAAACAAUUUCAGCUAC .((((....---........((((......----......)))--------------)...........((((........))))))))....((((-(((........))))))).... ( -16.10) >DroSec_CAF1 17371 98 + 1 AUCCAUGAC---AACCCCUAGAGCACCCAG----CUUCCAGUU--------------CCCUAUUUUUCCGUCCCCAUGGGCGGACGGAAAAACCUGA-AAUGGAAAACAAUUUCAGCUAC .(((((..(---(.......((((.....)----)))......--------------.....(((((((((((((...)).)))))))))))..)).-.)))))................ ( -25.90) >DroSim_CAF1 18195 98 + 1 AUCCAUGAC---AACCCCUCGAGCACCCAG----CUUCCAGUU--------------CCCUAUUUUUCCGUCCCCAUGGGCGGACGGAAAAACCUGA-AAUGGAAAACAAUUUCAGCUAC .(((((..(---(.......((((.....)----)))......--------------.....(((((((((((((...)).)))))))))))..)).-.)))))................ ( -25.90) >DroEre_CAF1 17890 97 + 1 AUCCAUGAC---AACCCCUCGAGCUCCCAG----CUUCCAACU--------------ACCUAUUUUUCCGAC-CCAUGGGCGGGCGGAAAAACCUGA-AAUGGAAAACAAUUUCAGCUAC .(((((..(---(.......((((.....)----)))......--------------.....((((((((.(-((......)))))))))))..)).-.)))))................ ( -23.40) >DroYak_CAF1 18162 112 + 1 AUCCAUGAC---AACCCCUCGAGCACCCAU----CUUCCAGCUCCCAGCAUACAGUUCCCUAUUUUUCCGUCCCCAUGGGCGGGCGGAAAAACCUGA-AAUGGAAAACAAUUUCAGCUAC .........---........((((......----......))))..(((.............(((((((((((((...)).)))))))))))..(((-(((........))))))))).. ( -27.10) >DroAna_CAF1 14886 105 + 1 UUCUGAAACUGGAGCUCCUGGGUCUCCUGAACUCCUGACUCCU--------------CGCAGCUUCUAGCUCCCCGCGGGCGGUG-GGAAAACCUGAAAAUGGAAAACAAUUUCAGCUAC ..........((((((....)).)))).....((((.(((.((--------------((((((.....)))....))))).))))-)))....((((((.((.....)).)))))).... ( -28.60) >consensus AUCCAUGAC___AACCCCUCGAGCACCCAG____CUUCCAGCU______________CCCUAUUUUUCCGUCCCCAUGGGCGGACGGAAAAACCUGA_AAUGGAAAACAAUUUCAGCUAC .....................(((......................................(((((((((((((...)).)))))))))))........(((((.....)))))))).. (-12.61 = -13.45 + 0.84)

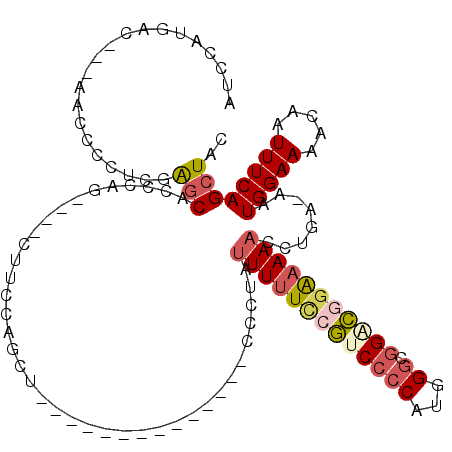

| Location | 14,667,649 – 14,667,747 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.84 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14667649 98 + 23771897 GUU--------------CCCUAUUUUUUCGUCCCCAUGGCCGGACUGGAAAACCUGAAAUGGAAAACAAUUUCAGCUACGCAAACACGGCGGGAAGGCCAACACAAAA----AAAA .((--------------(((...((((((((((........)))).)))))).(((((((........)))))))....((.......))))))).............----.... ( -21.30) >DroSec_CAF1 17404 95 + 1 GUU--------------CCCUAUUUUUCCGUCCCCAUGGGCGGACGGAAAAACCUGAAAUGGAAAACAAUUUCAGCUACGCAAACACGGCGGGAAGGCCAACACAAAA-------A .((--------------(((..(((((((((((((...)).))))))))))).(((((((........)))))))....((.......))))))).............-------. ( -30.80) >DroSim_CAF1 18228 96 + 1 GUU--------------CCCUAUUUUUCCGUCCCCAUGGGCGGACGGAAAAACCUGAAAUGGAAAACAAUUUCAGCUACGCAAACACGGCGGGAAGGCCAACACAAAA------AA .((--------------(((..(((((((((((((...)).))))))))))).(((((((........)))))))....((.......))))))).............------.. ( -30.80) >DroEre_CAF1 17923 101 + 1 ACU--------------ACCUAUUUUUCCGAC-CCAUGGGCGGGCGGAAAAACCUGAAAUGGAAAACAAUUUCAGCUACGCAAACACGGCGAGAAGGCCAACACAAAAUCACAAAA ...--------------.(((.((((((((.(-((......))))))))))).(((((((........)))))))((.(((.......))))).)))................... ( -25.40) >DroYak_CAF1 18195 107 + 1 GCUCCCAGCAUACAGUUCCCUAUUUUUCCGUCCCCAUGGGCGGGCGGAAAAACCUGAAAUGGAAAACAAUUUCAGCUACGCAAACACGGCGAGAAGGCCAACACAA---------A .......((.............(((((((((((((...)).))))))))))).(((((((........)))))))....))......(((......))).......---------. ( -30.00) >consensus GUU______________CCCUAUUUUUCCGUCCCCAUGGGCGGACGGAAAAACCUGAAAUGGAAAACAAUUUCAGCUACGCAAACACGGCGGGAAGGCCAACACAAAA______AA ......................(((((((((((........))))))))))).(((((((........)))))))............(((......)))................. (-22.62 = -22.78 + 0.16)


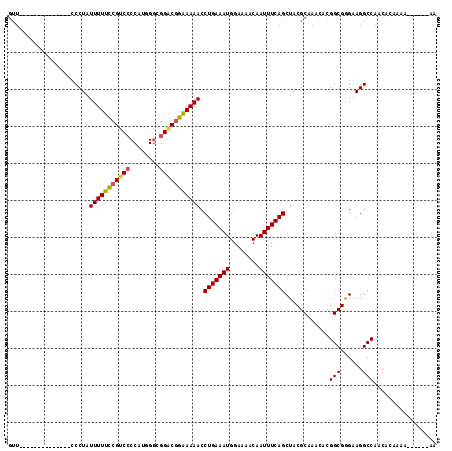
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:53 2006