
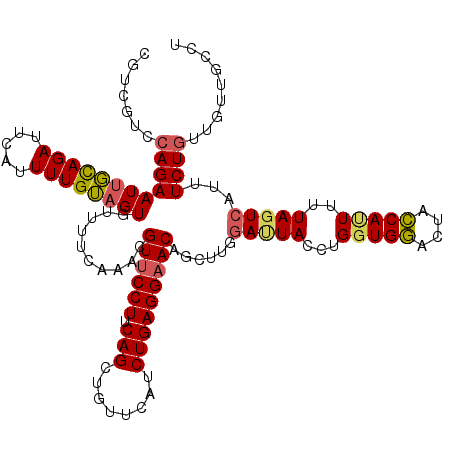
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,665,769 – 14,665,889 |
| Length | 120 |
| Max. P | 0.737124 |

| Location | 14,665,769 – 14,665,889 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -20.37 |
| Energy contribution | -20.66 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14665769 120 + 23771897 CGUCGUCCAGAAUUGCAGAUUCAUUUUGUAGUGUUUUCAAAUGUUCCUUCAGCUGUUCAUCUGAGGAACAGCUUGGACUACCUGGUGGACUACCAUUUUUAAUCAUUUCUGUUGUUGCCU .(((..((((.((((((((.....))))))))...(((((.(((((((.(((........))))))))))..)))))....))))..))).............................. ( -29.40) >DroSec_CAF1 15517 120 + 1 CGUCGUCCAGAAUUGCAGAUUCAUUUUGUAGUGUUUUCAAACGUUCCUUCAGCUGUUCAUCUGAGGAACAGCUUGGAUUACCUGGUGGACUACCACUUUUAGUCAUUUCUGUUGUUGCCU ...((.((((.((((((((.....))))))))............(((...((((((((.......)))))))).)))....))))))(((((.......)))))................ ( -29.30) >DroSim_CAF1 16332 120 + 1 CGUCGUCCAGAAUUGCAGAUUCAUUUUGUAGUGUUUUCAAACGUUCCUUCAGCUGUUCAUCUGAGGAACAGCUUAGACUACCUGGUGGACUACCAUUUAUAGUCAUUUCUUUUGUUGCCU (((........((((((((.....))))))))........))).......((((((((.......))))))))..(((((...(((((....)))))..)))))................ ( -27.19) >DroEre_CAF1 16028 120 + 1 CGUCAUCCAGAAUCUUAGACUCAUUUUGAAGUGCUUUAAUACGUUCCUUCAGCUGUUCAUCUGAGGAACAGCUUUGAUUACUUGUUGAACUUUCAUUUUUAGUCAUUUCUGGUGAUGCCU (((((.((((((.....((((.....(((((.(.(((((((.((......((((((((.......))))))))......)).))))))))))))).....))))..)))))))))))... ( -34.30) >DroYak_CAF1 16288 105 + 1 CGUCAUCCAGAAUUUUAGAUUCGUUUUGAAGUGCUUUAAUACGUUCCUCCAGCUGUUUAUCUGAGGCACAGUUUUCAUUACU---------------UUUAGUAAUUUCUGUUGAUGCCU (((((..(((((..(((((...((..((((((((((((..(((((.....))).)).....))))))))....))))..)).---------------)))))....))))).)))))... ( -20.20) >consensus CGUCGUCCAGAAUUGCAGAUUCAUUUUGUAGUGUUUUCAAACGUUCCUUCAGCUGUUCAUCUGAGGAACAGCUUGGAUUACCUGGUGGACUACCAUUUUUAGUCAUUUCUGUUGUUGCCU .......((((((((((((.....))))))))..........((((((.(((........)))))))))......(((((...(((((....)))))..)))))...))))......... (-20.37 = -20.66 + 0.29)


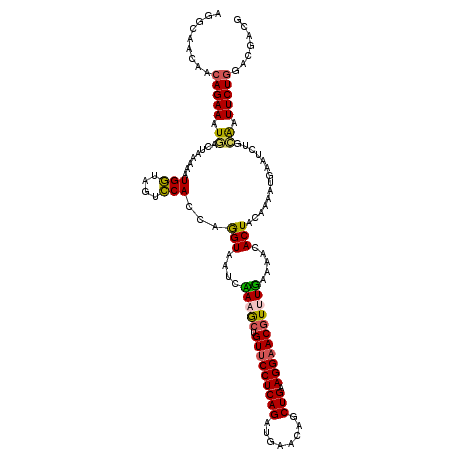
| Location | 14,665,769 – 14,665,889 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -26.79 |
| Consensus MFE | -16.03 |
| Energy contribution | -15.78 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14665769 120 - 23771897 AGGCAACAACAGAAAUGAUUAAAAAUGGUAGUCCACCAGGUAGUCCAAGCUGUUCCUCAGAUGAACAGCUGAAGGAACAUUUGAAAACACUACAAAAUGAAUCUGCAAUUCUGGACGACG .(....)..(((((.((.......((.(((((....(((((..(((.((((((((.......))))))))...)))..))))).....)))))...)).......)).)))))....... ( -30.24) >DroSec_CAF1 15517 120 - 1 AGGCAACAACAGAAAUGACUAAAAGUGGUAGUCCACCAGGUAAUCCAAGCUGUUCCUCAGAUGAACAGCUGAAGGAACGUUUGAAAACACUACAAAAUGAAUCUGCAAUUCUGGACGACG .(....)..(((((.((.......((((....))))(((((..(((.((((((((.......))))))))...))).(((((............))))).))))))).)))))....... ( -30.80) >DroSim_CAF1 16332 120 - 1 AGGCAACAAAAGAAAUGACUAUAAAUGGUAGUCCACCAGGUAGUCUAAGCUGUUCCUCAGAUGAACAGCUGAAGGAACGUUUGAAAACACUACAAAAUGAAUCUGCAAUUCUGGACGACG .(....).........((((((.....))))))..(((((((((.(((((.(((((((((........))).))))))))))).....)))))....((......))...))))...... ( -28.00) >DroEre_CAF1 16028 120 - 1 AGGCAUCACCAGAAAUGACUAAAAAUGAAAGUUCAACAAGUAAUCAAAGCUGUUCCUCAGAUGAACAGCUGAAGGAACGUAUUAAAGCACUUCAAAAUGAGUCUAAGAUUCUGGAUGACG .....(((((((((..((((.....((((.((((.............((((((((.......))))))))....))))((......))..)))).....)))).....)))))).))).. ( -27.93) >DroYak_CAF1 16288 105 - 1 AGGCAUCAACAGAAAUUACUAAA---------------AGUAAUGAAAACUGUGCCUCAGAUAAACAGCUGGAGGAACGUAUUAAAGCACUUCAAAACGAAUCUAAAAUUCUGGAUGACG .(.((((..((((((((((....---------------.)))))(((....((.((((((........)).)))).))((......))..)))...............))))))))).). ( -17.00) >consensus AGGCAACAACAGAAAUGACUAAAAAUGGUAGUCCACCAGGUAAUCAAAGCUGUUCCUCAGAUGAACAGCUGAAGGAACGUUUGAAAACACUACAAAAUGAAUCUGCAAUUCUGGACGACG .........(((((.((........(((....)))...(((....(((((.(((((((((........))).))))))))))).....)))..............)).)))))....... (-16.03 = -15.78 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:51 2006