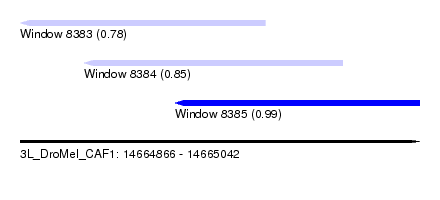
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,664,866 – 14,665,042 |
| Length | 176 |
| Max. P | 0.993126 |

| Location | 14,664,866 – 14,664,974 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.80 |
| Mean single sequence MFE | -21.37 |
| Consensus MFE | -10.73 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14664866 108 - 23771897 UCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAGAUAAUUCGUAAAUACAAAUUAAACAAAAAAAUGUUAAUGUUAAAUUAAACAAAUGUUGUAUUUGAAAAUC ((.((((((((..((.......))..)))))))).))....(((..(((((((((....((((......))))..((((......))))....)))))))))..))). ( -23.30) >DroSec_CAF1 14659 81 - 1 UCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAUGUAUUUCGUGGAUA-------------------------GUAUAUUAAA--AAUGUUGUAUUCGAAAAAC ((.((((((((..((.......))..)))))))).))....(((((...(((-------------------------((((......--.)))))))...)))))... ( -19.20) >DroSim_CAF1 15473 82 - 1 UCCUGGUUGGGGGCAUCGAUUUUGACCGCAAUCGGGAUGUACUUAAUGGGAA-------------------------GUAUAUUAAAU-AAUGUUGUAUUCGGAAAAC (((((((((.((.((.......)).)).)))))))))(((((((......))-------------------------)))))......-................... ( -21.60) >consensus UCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAUGUAAUUCGUGGAUA_________________________GUAUAUUAAA__AAUGUUGUAUUCGAAAAAC ((.((((((((..((.......))..)))))))).))....................................................................... (-10.73 = -11.07 + 0.33)


| Location | 14,664,894 – 14,665,008 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.48 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -16.02 |
| Energy contribution | -15.90 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14664894 114 - 23771897 -CCCAAGCCGAAGCCAACACGAGAAUGGUCUGGUGUCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAGAUAAUUCGUAAAUACAAAUUAAACAAAAAAAUGUUAAUGUUA -(((............((((.(((....))).))))...(((((((..((.......))..))))))))))...(((((.((....)).)))))((((......))))....... ( -26.50) >DroSec_CAF1 14685 89 - 1 -CCCAAGCCGAAGCCAACACGAAAAUGGUCUGGGGUCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAUGUAUUUCGUGGAUA-------------------------GUA -((((..(((...(......)....)))..))))(((.((((((((..((.......))..)))))))).)))..............-------------------------... ( -25.50) >DroSim_CAF1 15500 89 - 1 -CCCAAGCCGAAGCCAACACGAAAAUGGUCUGGGGUCCUGGUUGGGGGCAUCGAUUUUGACCGCAAUCGGGAUGUACUUAAUGGGAA-------------------------GUA -((((..((.(..(((.........)))..).))((((((((((.((.((.......)).)).))))))))))........))))..-------------------------... ( -31.10) >DroEre_CAF1 15157 90 - 1 CCGCAAGGAAAAGCCAAAAAGAAAGUGGAGUGGGGUUUUGGUUGGGGGUAUCCAUUUUGAACCCGAACGCCUAAAGCUUCCUUUAUG-------------------------GUC ((....))....((((.((((.((((....((((((((.((((.(((((....))))).)))).)))).))))..)))).)))).))-------------------------)). ( -29.80) >consensus _CCCAAGCCGAAGCCAACACGAAAAUGGUCUGGGGUCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAUAUACUUCGUGGAUA_________________________GUA .(((.....((..(((.((......))...)))..))..(((((((..((.......))..))))))))))............................................ (-16.02 = -15.90 + -0.12)



| Location | 14,664,934 – 14,665,042 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.61 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -12.03 |
| Energy contribution | -12.11 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.43 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14664934 108 - 23771897 AAAGAAGAAACAAAAAAAUAAAUCUAAGCGUAAG------------CCCAAGCCGAAGCCAACACGAGAAUGGUCUGGUGUCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAGAU .....................((((..((....)------------).....(((((((((((((.(((....))).))))..)))))(((..((.......))..)))..)))).)))) ( -29.40) >DroSec_CAF1 14700 96 - 1 AAAGAAAAAAAAGAAAAAUAAA------------------------CCCAAGCCGAAGCCAACACGAAAAUGGUCUGGGGUCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAUGU ......................------------------------((((..(((...(......)....)))..))))(((.((((((((..((.......))..)))))))).))).. ( -26.00) >DroSim_CAF1 15515 96 - 1 AAAGAAGAAAAAAAAAAAUAAA------------------------CCCAAGCCGAAGCCAACACGAAAAUGGUCUGGGGUCCUGGUUGGGGGCAUCGAUUUUGACCGCAAUCGGGAUGU ......................------------------------((((..(((...(......)....)))..))))((((((((((.((.((.......)).)).)))))))))).. ( -28.60) >DroEre_CAF1 15172 114 - 1 AAAGGUAGUACCAAAAUAUAAACCCACACGUAAGGUUAG------CCGCAAGGAAAAGCCAAAAAGAAAGUGGAGUGGGGUUUUGGUUGGGGGUAUCCAUUUUGAACCCGAACGCCUAAA ..((((....(((((((....((((((......((((..------((....))...)))).........)))).))...)))))))(((((..((.......))..)))))..))))... ( -32.36) >DroYak_CAF1 15483 120 - 1 AAAGAAAAAACCUGUAAAGAAACCUAAGCGUAAGCCCAAGCCCAAACCCAAGCCCAAGCCAACACGAAAACGUUAUAAGGCUAUCGUUGGGGGCAUCGAUUUUGAACCCGAUGGCCACAA ............(((............((....))................(((..((((...(((....))).....))))...((((((..((.......))..))))))))).))). ( -23.30) >consensus AAAGAAAAAACAAAAAAAUAAA_C_A__CGUAAG____________CCCAAGCCGAAGCCAACACGAAAAUGGUCUGGGGUCAUGGUUGGGGGCAUCGAUUUUGAACCCAAUCGGGAUAU ..............................................(((........(((..................)))...(((((((..((.......))..)))))))))).... (-12.03 = -12.11 + 0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:49 2006