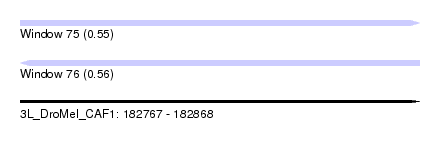
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 182,767 – 182,868 |
| Length | 101 |
| Max. P | 0.564970 |

| Location | 182,767 – 182,868 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.98 |
| Mean single sequence MFE | -46.35 |
| Consensus MFE | -32.26 |
| Energy contribution | -32.23 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
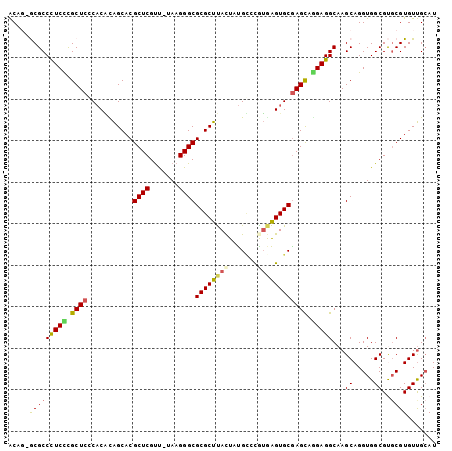
>3L_DroMel_CAF1 182767 101 + 23771897 ACAG-GCGCCCUCCCGCUCGCACAUAGCACGCUCGUU-UAAGGGCGCGCUUACUAUGCGCGUGAGUGCAAGCAGGAGGCAAGCAGGUGGCGUUCGUGUUGCAU ....-((..(((((.(((.((((....(((((((...-...))))((((.......))))))).)))).))).)))))...))..(..(((....)))..).. ( -43.60) >DroPse_CAF1 2175 102 + 1 ACUC-ACGCCUUCUCGCUCCCACACAGCACGCUCGUUUUAAGGGCGCGCUUACUCUGUCUAUGAGUGCGAGCGGGAGGCAAGCAGGUGGCGCGCGUGUUGCAU ....-..((((((.(((((......(((.(((((.......))))).)))(((((.......))))).)))))))))))......(..(((....)))..).. ( -40.80) >DroEre_CAF1 13589 101 + 1 ACAG-GCGCCCUCCCGCUCGCACGCAGCACGCUCGUU-UAAGGGCGCGCUUACUAUGCGCGUGAGUGCGAGUAAGAGGCAAGCAGGUGGCGUGCGUGUUGCAU (((.-((((((.((.(((.((...(.((.(((((...-...))))).))(((((.(((((....)))))))))))..)).))).)).)).)))).)))..... ( -47.30) >DroYak_CAF1 3532 101 + 1 ACAG-GCGCCCUCCUGCUCGCACGCAGCACGCUCGUU-UAAGGGCGCGCUUACUGUGCGCGUAAGUGCGAGCAGGAGGCAAGCAGGUGGCGUGCGUGUUGCAU (((.-((((((((((((((((((...((..((((...-...))))((((.....))))))....))))))))))))))...((.....)))))).)))..... ( -55.60) >DroAna_CAF1 12435 102 + 1 GCCGUGCGCCCUCUCGCUCCCUCUUGGCACGCUCGCC-UAAGGGCGCGCUCAUUGCGGCCGAAGGUGCGAGCAAGAGGCAAGCAGGUGGCGUGCGUGUCGGAU ((((..((((.(((.(((.(((((((.(.(((..(((-(..((.((((.....)))).))..))))))).))))))))..)))))).))))..)).))..... ( -50.00) >DroPer_CAF1 6094 102 + 1 ACUC-ACGCCUUCUCGCUCCCACACAGCACGCUCGUUUUAAGGGCGCGCUUACUCUGUCUAUGAGUGCGAGCGGGAGGCAAGCAGGUGGCGCGCGUGUUGCAU ....-..((((((.(((((......(((.(((((.......))))).)))(((((.......))))).)))))))))))......(..(((....)))..).. ( -40.80) >consensus ACAG_GCGCCCUCCCGCUCCCACACAGCACGCUCGUU_UAAGGGCGCGCUUACUAUGCCCGUGAGUGCGAGCAGGAGGCAAGCAGGUGGCGUGCGUGUUGCAU .....(((((((((.((((...........((((.......))))((((((((.......)))))))))))).)))))...((.....))..))))....... (-32.26 = -32.23 + -0.02)

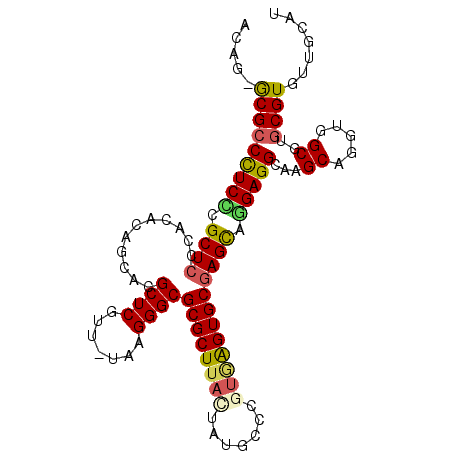
| Location | 182,767 – 182,868 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.98 |
| Mean single sequence MFE | -43.63 |
| Consensus MFE | -29.97 |
| Energy contribution | -30.61 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
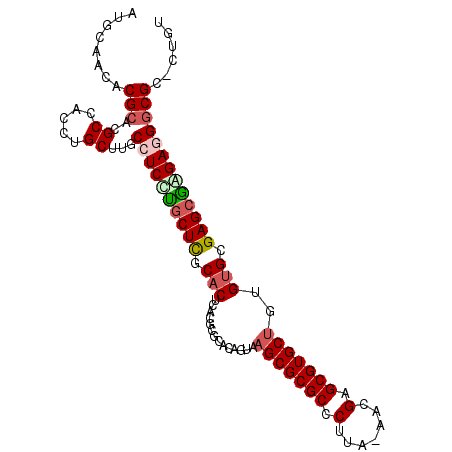
>3L_DroMel_CAF1 182767 101 - 23771897 AUGCAACACGAACGCCACCUGCUUGCCUCCUGCUUGCACUCACGCGCAUAGUAAGCGCGCCCUUA-AACGAGCGUGCUAUGUGCGAGCGGGAGGGCGC-CUGU ..((.........((.....))...((((((((((((((....((.....)).(((((((.(...-...).)))))))..))))))))))))))))..-.... ( -41.90) >DroPse_CAF1 2175 102 - 1 AUGCAACACGCGCGCCACCUGCUUGCCUCCCGCUCGCACUCAUAGACAGAGUAAGCGCGCCCUUAAAACGAGCGUGCUGUGUGGGAGCGAGAAGGCGU-GAGU .......((.((((((.....(((((.((((((.(..((((.......)))).(((((((.(.......).)))))))).)))))))))))..)))))-).)) ( -43.00) >DroEre_CAF1 13589 101 - 1 AUGCAACACGCACGCCACCUGCUUGCCUCUUACUCGCACUCACGCGCAUAGUAAGCGCGCCCUUA-AACGAGCGUGCUGCGUGCGAGCGGGAGGGCGC-CUGU .(((.....)))((((.((((((((((...(((((((......)))...))))(((((((.(...-...).)))))))..).)))))))))..)))).-.... ( -41.10) >DroYak_CAF1 3532 101 - 1 AUGCAACACGCACGCCACCUGCUUGCCUCCUGCUCGCACUUACGCGCACAGUAAGCGCGCCCUUA-AACGAGCGUGCUGCGUGCGAGCAGGAGGGCGC-CUGU ..(((...(((..((.....))...((((((((((((((.(((.......)))(((((((.(...-...).)))))))..))))))))))))))))).-.))) ( -47.70) >DroAna_CAF1 12435 102 - 1 AUCCGACACGCACGCCACCUGCUUGCCUCUUGCUCGCACCUUCGGCCGCAAUGAGCGCGCCCUUA-GGCGAGCGUGCCAAGAGGGAGCGAGAGGGCGCACGGC ........((..((((..((((((.((((((((.(((.(((..((((((.....))).)))...)-))...))).).))))))))))).))..))))..)).. ( -45.10) >DroPer_CAF1 6094 102 - 1 AUGCAACACGCGCGCCACCUGCUUGCCUCCCGCUCGCACUCAUAGACAGAGUAAGCGCGCCCUUAAAACGAGCGUGCUGUGUGGGAGCGAGAAGGCGU-GAGU .......((.((((((.....(((((.((((((.(..((((.......)))).(((((((.(.......).)))))))).)))))))))))..)))))-).)) ( -43.00) >consensus AUGCAACACGCACGCCACCUGCUUGCCUCCUGCUCGCACUCACGCGCACAGUAAGCGCGCCCUUA_AACGAGCGUGCUGUGUGCGAGCGAGAGGGCGC_CUGU ........(((..((.....))...((((((((((.(((..............(((((((.(.......).)))))))..))).)))))))))))))...... (-29.97 = -30.61 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:08 2006