| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,651,696 – 14,651,792 |
| Length | 96 |
| Max. P | 0.730736 |

| Location | 14,651,696 – 14,651,792 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -20.79 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
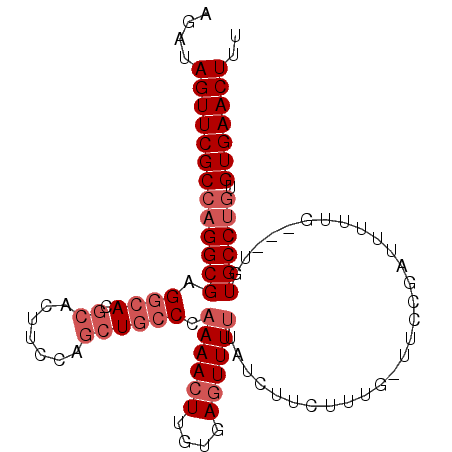
>3L_DroMel_CAF1 14651696 96 + 23771897 AAAGUUCACACAGGCACA---CAAAAAUCGGAA-AAAAGAAGAUAAAAACUCACAAGUUUUGGGCAGCUGGAAGUGCGUGCCUCGCCUGGCGAACUAUCU ..(((((.(.(((((...---............-...........((((((....))))))(((((((.......)).))))).)))))).))))).... ( -26.90) >DroSec_CAF1 1531 96 + 1 AAAGUUCACACAGGCACA---CAAAAAUCGGGA-CAAAGAAGAUAAAAACUCACAAGUUUUGGGCAGCUGGAAGUGCGUGCCUCGCCUGGCGAACUAUCU ..(((((.(.(((((...---............-...........((((((....))))))(((((((.......)).))))).)))))).))))).... ( -26.90) >DroSim_CAF1 1524 96 + 1 AAAGUUCACACAGGCACA---CAAAAAUCGGAA-CAAAGAAGAUAAAAACUCACAAGUUUUGGGCAGCUGGAAGUGCGUGCCUCGCCUGGCGAACUAUCU ..(((((.(.(((((...---............-...........((((((....))))))(((((((.......)).))))).)))))).))))).... ( -26.90) >DroEre_CAF1 1868 96 + 1 UAAGUUCACACAGGCACU---CAACAAUCGGAA-CAAAAAAGAUAAAAACUCACAAGUUUUGGGCAGCUGCAAGUGCGUGCCUCGCCUGGCGAACUCGCU ..(((((.(.(((((...---............-...........((((((....))))))(((((((.......)).))))).)))))).))))).... ( -26.30) >DroYak_CAF1 1520 96 + 1 UAAGUUCACACGGGCACA---CAAAAAUCGGCAAAAAAAAAGAUA-AAACUCAAAAGUUUUGGGCAGCUGGAAAUGCGUGCCUCGCCUGGCGAACUCGCU ..(((((.(.(((((...---......................((-(((((....)))))))((((((.......)).))))..)))))).))))).... ( -26.00) >DroAna_CAF1 1709 97 + 1 AAAGUUCACAGAGGCAUAUAGCAAGAAGCCAUA-AAAAAACGACAAAAAGUCACAAGUUUUUGGGA--CAGAAGUGCGUGUCUCGCCUGGCGAACUAGUU ..(((((.(..((((.(((.((.....)).)))-.(((((((((.....)))....))))))((((--((........)))))))))).).))))).... ( -23.70) >consensus AAAGUUCACACAGGCACA___CAAAAAUCGGAA_AAAAAAAGAUAAAAACUCACAAGUUUUGGGCAGCUGGAAGUGCGUGCCUCGCCUGGCGAACUAGCU ..(((((.(.(((((..............................((((((....))))))(((((((.......)).))))).)))))).))))).... (-20.79 = -21.68 + 0.89)


| Location | 14,651,696 – 14,651,792 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 86.71 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -21.63 |
| Energy contribution | -22.97 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14651696 96 - 23771897 AGAUAGUUCGCCAGGCGAGGCACGCACUUCCAGCUGCCCAAAACUUGUGAGUUUUUAUCUUCUUUU-UUCCGAUUUUUG---UGUGCCUGUGUGAACUUU ....(((((((((((((.((((.((.......))))))(((((.(((.(((...............-)))))).)))))---..)))))).))))))).. ( -29.46) >DroSec_CAF1 1531 96 - 1 AGAUAGUUCGCCAGGCGAGGCACGCACUUCCAGCUGCCCAAAACUUGUGAGUUUUUAUCUUCUUUG-UCCCGAUUUUUG---UGUGCCUGUGUGAACUUU ....(((((((((((((.((((.((.......)))))))((((((....))))))...........-............---...))))).))))))).. ( -28.20) >DroSim_CAF1 1524 96 - 1 AGAUAGUUCGCCAGGCGAGGCACGCACUUCCAGCUGCCCAAAACUUGUGAGUUUUUAUCUUCUUUG-UUCCGAUUUUUG---UGUGCCUGUGUGAACUUU ....(((((((((((((.((((.((.......))))))(((((.(((.(((...............-)))))).)))))---..)))))).))))))).. ( -28.76) >DroEre_CAF1 1868 96 - 1 AGCGAGUUCGCCAGGCGAGGCACGCACUUGCAGCUGCCCAAAACUUGUGAGUUUUUAUCUUUUUUG-UUCCGAUUGUUG---AGUGCCUGUGUGAACUUA ...((((((((((((((.((((.((.......)))))))((((((....))))))...........-............---...))))).)))))))). ( -30.20) >DroYak_CAF1 1520 96 - 1 AGCGAGUUCGCCAGGCGAGGCACGCAUUUCCAGCUGCCCAAAACUUUUGAGUUU-UAUCUUUUUUUUUGCCGAUUUUUG---UGUGCCCGUGUGAACUUA ...((((((((...(((.((((((((..((..((.....((((((....)))))-)............)).))....))---))))))))))))))))). ( -27.43) >DroAna_CAF1 1709 97 - 1 AACUAGUUCGCCAGGCGAGACACGCACUUCUG--UCCCAAAAACUUGUGACUUUUUGUCGUUUUUU-UAUGGCUUCUUGCUAUAUGCCUCUGUGAACUUU ....(((((((.((((((((.(((.((....(--((.(((....))).))).....))))).))))-((((((.....)))))).))))..))))))).. ( -27.10) >consensus AGAUAGUUCGCCAGGCGAGGCACGCACUUCCAGCUGCCCAAAACUUGUGAGUUUUUAUCUUCUUUG_UUCCGAUUUUUG___UGUGCCUGUGUGAACUUU ....(((((((((((((.((((.((.......)))))).((((((....)))))).............................)))))).))))))).. (-21.63 = -22.97 + 1.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:37 2006