
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,627,763 – 14,627,885 |
| Length | 122 |
| Max. P | 0.663096 |

| Location | 14,627,763 – 14,627,866 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -15.27 |
| Energy contribution | -16.18 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
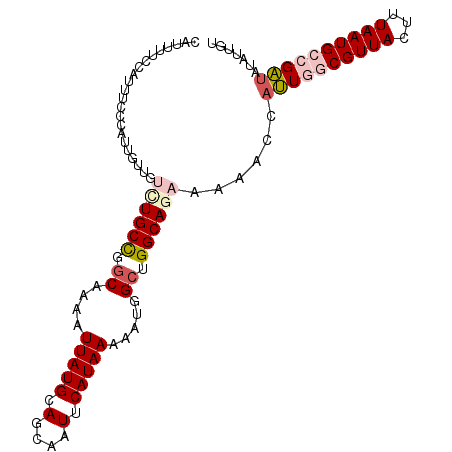
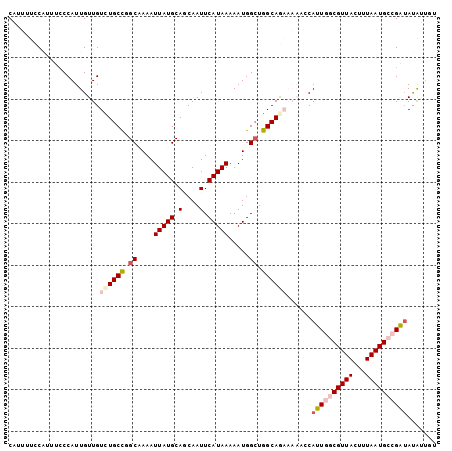
>3L_DroMel_CAF1 14627763 103 + 23771897 CAUUUUCCAUUUCCCAUUGUUGUCUGCCUGCGAAAUUAUGCAGCAAUUCAUAAAAAUGGCUGGCAGAAAAACCAUUGGCGUUACUUUAAUGCCGAUAUGUUUU ......................((((((.((.(..(((((.(....).)))))...).)).))))))(((((.((((((((((...))))))))))..))))) ( -26.40) >DroPse_CAF1 30822 93 + 1 CUGG-UAC---ACCCAUUGUUGUCUGCUGGCCAAAUUAUGCAUCAAUUCAUAAAAAUGGCUGGCAA------CAGUCCCGUUACUUUAAUGCCGUUACAGUGU ..((-...---.))(((((((((.(((..((((..(((((........)))))...))))..))))------))....(((((...))))).....)))))). ( -21.40) >DroSec_CAF1 26645 103 + 1 CAUUUUCCAUUUCCCAUUGUUGUCUGCCUGCAAAAUUAUGCAGCAAUUCAUAAAAAUGGCUGGCAGAAAAACCAUUGGCGUUACUUUAAUGCCGAUAUAUUUU ..................(((.((((((.((....(((((.(....).))))).....)).))))))..))).((((((((((...))))))))))....... ( -25.60) >DroSim_CAF1 22555 102 + 1 CAUUUUCCAUUUCCCAUUGUUGUUUGCCUGCAAAAUUAUGCAGCAAUUCAUAAAAAUGGAUGGCAGAAAACCCAUUGGCGUUACUUUAAUGCCGA-AUAUNNN ..((((((((...(((((..((.((((.((((......))))))))..))....))))))))).))))......(((((((((...)))))))))-....... ( -25.70) >DroEre_CAF1 29969 103 + 1 CAUUUUCCAUUUGCCAUUGUUGUCUGCCGGCAAAAUUAUGCAGCAAUUCAUAAAAAUGGCUGGCAGAAAAACCAUUGGCGUUACUUUAAUGCCGAUAUGUUAU ..................(((.(((((((((....(((((.(....).))))).....)))))))))..))).((((((((((...))))))))))....... ( -29.50) >DroPer_CAF1 30822 93 + 1 CUGG-UAC---ACCCAUUGUUGUCUGCUGGCCAAAUUAUGCAUCAAUUCAUAAAAAUGGCUGGCAA------CAGUCCCGUUACUUUAAUGCCGUUACAGUGU ..((-...---.))(((((((((.(((..((((..(((((........)))))...))))..))))------))....(((((...))))).....)))))). ( -21.40) >consensus CAUUUUCCAUUUCCCAUUGUUGUCUGCCGGCAAAAUUAUGCAGCAAUUCAUAAAAAUGGCUGGCAGAAAAACCAUUGGCGUUACUUUAAUGCCGAUAUAUUGU ......................((((((.((....(((((........))))).....)).))))))......((((((((((...))))))))))....... (-15.27 = -16.18 + 0.92)

| Location | 14,627,787 – 14,627,885 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -10.75 |
| Energy contribution | -11.58 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
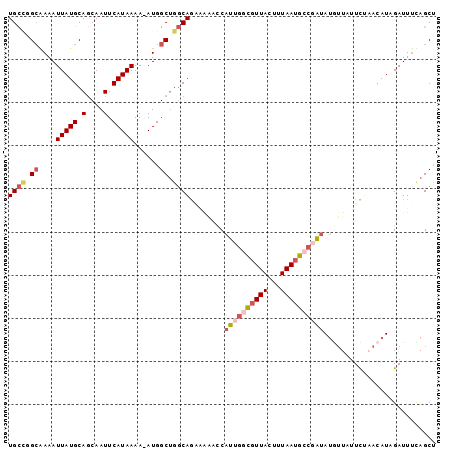
>3L_DroMel_CAF1 14627787 98 + 23771897 UGCCUGCGAAAUUAUGCAGCAAUUCAUAAAA-AUGGCUGGCAGAAAAACCAUUGGCGUUACUUUAAUGCCGAUAUGUUUUUCUAACAUUGAUUUUAGCU .(((.((.(..(((((.(....).)))))..-.).)).)))((((((((.((((((((((...))))))))))..))))))))................ ( -27.10) >DroGri_CAF1 34867 97 + 1 UGU-GGAGAAAUUAUGCAGCAAUUCAUAAAAAAUGGCAACCAAU-GGCCCAGCGAUAUUACUUUAAGACCCUAAUGUUAUUUUCACAUAGAUUUCAGCA (((-(((((..(((((.(....).))))).....(((.......-.)))....(((((((...........))))))).))))))))............ ( -13.10) >DroSec_CAF1 26669 98 + 1 UGCCUGCAAAAUUAUGCAGCAAUUCAUAAAA-AUGGCUGGCAGAAAAACCAUUGGCGUUACUUUAAUGCCGAUAUAUUUUUCUAACAUCGAUUUUAGCU (((.((((......))))))).......(((-((.(.((..(((((((..((((((((((...))))))))))...)))))))..)).).))))).... ( -26.50) >DroEre_CAF1 29993 98 + 1 UGCCGGCAAAAUUAUGCAGCAAUUCAUAAAA-AUGGCUGGCAGAAAAACCAUUGGCGUUACUUUAAUGCCGAUAUGUUAUUCAAACAUAGAUUUCAGCU (((((((....(((((.(....).)))))..-...)))))))((((.....(((((((((...)))))))))((((((.....))))))..)))).... ( -28.60) >DroYak_CAF1 30482 98 + 1 UGCCUGCAAAAUUAUGCAGCAAUUCAUAAAA-AUGGCUGGCAGAAAAUCCAUUGGCGUUACUUUAAUGCCUAUAUGUUAGUCUAACAUAGAUUUUAGCU ((((.((....(((((.(....).)))))..-...)).)))).((((((....(((((((...)))))))..(((((((...))))))))))))).... ( -24.10) >DroPer_CAF1 30842 88 + 1 UGCUGGCCAAAUUAUGCAUCAAUUCAUAAAA-AUGGCUGGCAA------CAGUCCCGUUACUUUAAUGCCGUUACAGUGUUU----AUCGAUUGCAGCC (((..((((..(((((........)))))..-.))))..))).------(((((....((((.((((...)))).))))...----...)))))..... ( -20.30) >consensus UGCCGGCAAAAUUAUGCAGCAAUUCAUAAAA_AUGGCUGGCAGAAAAACCAUUGGCGUUACUUUAAUGCCGAUAUGUUAUUCUAACAUAGAUUUCAGCU ((((.((....(((((........)))))......)).))))........((((((((((...)))))))))).......................... (-10.75 = -11.58 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:31 2006