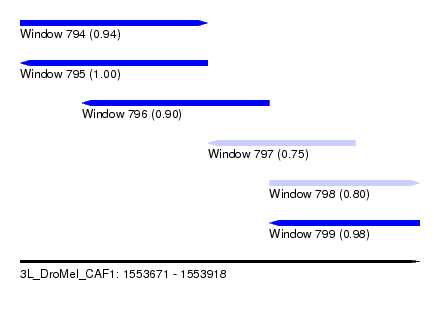
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,553,671 – 1,553,918 |
| Length | 247 |
| Max. P | 0.995613 |

| Location | 1,553,671 – 1,553,787 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -22.96 |
| Energy contribution | -23.52 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1553671 116 + 23771897 UUCUGCCUCUUCUUUUGGCCAUUUACUUCUGGCUUUCGGUGUCGCCGAGAUUCGUAAGCAAAUUUCAAGCGUUUAACAUUUAACGCUU-UGCGUUUUGCUCACAAAUUCGAGUUCAA ......(((........((((........))))((((((.....))))))...((.((((((....(((((((........)))))))-.....)))))).))......)))..... ( -28.80) >DroSec_CAF1 11384 115 + 1 CUCUUCC-CUUCUUUUGGCCAUUUACUUCUGGCUUUCGGUGUCGCCGAGAUUCGUAAGCAAAUUUCAAGCGUUUAACAUUUAACGCUU-UGAGUUUUGCUCACAAAUUCGAGUUCAA (((....-.........((((........))))((((((.....))))))...((.((((((.((((((((((........))))).)-)))).)))))).))......)))..... ( -31.70) >DroSim_CAF1 11485 115 + 1 CUCUUCC-CUUCUUUUGGCCAUUUACUUCUGGCUUUCGGUGUCGCCGAGAUUCGUAAGCAAAUUUCAAGCGUUUAACAUUUAACGCUU-UGCGUUUUGCUCACAAAUUCGAGUUCAA (((....-.........((((........))))((((((.....))))))...((.((((((....(((((((........)))))))-.....)))))).))......)))..... ( -28.50) >DroYak_CAF1 12240 109 + 1 --------CUGCUUUUGGCCAUUUACUUCUGGCUUUCAGUGUCGUCGAGAUUCGUAAGCAAAUUUCAAGCAUUUAACAUUUAACGCUCUCGCCUUUUGCUCACAAAUUCGAGUUCAA --------..(((...(((((........)))))...)))....(((((.((.((.((((((.....(((.((........)).))).......)))))).)))).)))))...... ( -19.70) >consensus CUCUUCC_CUUCUUUUGGCCAUUUACUUCUGGCUUUCGGUGUCGCCGAGAUUCGUAAGCAAAUUUCAAGCGUUUAACAUUUAACGCUU_UGCGUUUUGCUCACAAAUUCGAGUUCAA .................((((........))))((((((.....))))))...((.((((((....(((((((........)))))))......)))))).)).............. (-22.96 = -23.52 + 0.56)

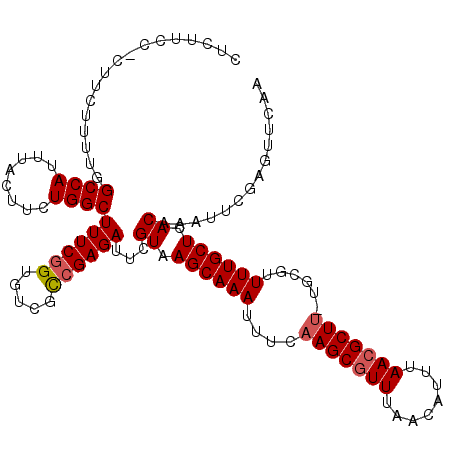
| Location | 1,553,671 – 1,553,787 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -30.64 |
| Energy contribution | -30.07 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1553671 116 - 23771897 UUGAACUCGAAUUUGUGAGCAAAACGCA-AAGCGUUAAAUGUUAAACGCUUGAAAUUUGCUUACGAAUCUCGGCGACACCGAAAGCCAGAAGUAAAUGGCCAAAAGAAGAGGCAGAA .....(((((.((((((((((((.....-(((((((........)))))))....))))))))))))))((((.....))))..((((........))))........)))...... ( -34.80) >DroSec_CAF1 11384 115 - 1 UUGAACUCGAAUUUGUGAGCAAAACUCA-AAGCGUUAAAUGUUAAACGCUUGAAAUUUGCUUACGAAUCUCGGCGACACCGAAAGCCAGAAGUAAAUGGCCAAAAGAAG-GGAAGAG .....(((...((((((((((((..(((-(.(((((........)))))))))..))))))))))))((((((.....))....((((........))))........)-))).))) ( -35.10) >DroSim_CAF1 11485 115 - 1 UUGAACUCGAAUUUGUGAGCAAAACGCA-AAGCGUUAAAUGUUAAACGCUUGAAAUUUGCUUACGAAUCUCGGCGACACCGAAAGCCAGAAGUAAAUGGCCAAAAGAAG-GGAAGAG .....(((...((((((((((((.....-(((((((........)))))))....))))))))))))((((((.....))....((((........))))........)-))).))) ( -33.80) >DroYak_CAF1 12240 109 - 1 UUGAACUCGAAUUUGUGAGCAAAAGGCGAGAGCGUUAAAUGUUAAAUGCUUGAAAUUUGCUUACGAAUCUCGACGACACUGAAAGCCAGAAGUAAAUGGCCAAAAGCAG-------- ......(((((((((((((((((......(((((((........)))))))....))))))))))))).)))).....(((...((((........))))......)))-------- ( -30.80) >consensus UUGAACUCGAAUUUGUGAGCAAAACGCA_AAGCGUUAAAUGUUAAACGCUUGAAAUUUGCUUACGAAUCUCGGCGACACCGAAAGCCAGAAGUAAAUGGCCAAAAGAAG_GGAAGAG (((...(((((((((((((((((......(((((((........)))))))....))))))))))))).))))...........((((........))))))).............. (-30.64 = -30.07 + -0.56)

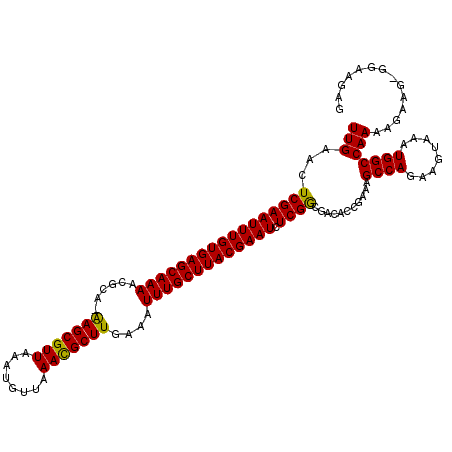
| Location | 1,553,709 – 1,553,825 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -12.97 |
| Energy contribution | -15.94 |
| Covariance contribution | 2.98 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1553709 116 - 23771897 GUCACGAACUCGUG-CAAAAGUU-AAUAAUACUCGCAUUGUUGAACUCGAAUUUGUGAGCA-AAACGCA-AAGCGUUAAAUGUUAAACGCUUGAAAUUUGCUUACGAAUCUCGGCGACAC ((((((....)))(-(.....((-((((((......))))))))...((((((((((((((-((.....-(((((((........)))))))....))))))))))))).)))))))).. ( -36.00) >DroPse_CAF1 12308 106 - 1 GUCACGAGCUCGGGGCCAACGCUGAAUCCCAAUCAUGAUGAU------GGCUUUUCAGGCGCACACACA-AAA-----GGCGAUAAACGAAUGA-AUUUGCGUACCAAUCUCG-CGACAC (((.((((....((.......(((((..(((.((.....)))------))...)))))(((((....((-...-----(........)...)).-...))))).))...))))-.))).. ( -25.80) >DroSec_CAF1 11421 116 - 1 GUCACGAACUUGUG-CAAAAGUU-AAUAAUACUCGCAUUGUUGAACUCGAAUUUGUGAGCA-AAACUCA-AAGCGUUAAAUGUUAAACGCUUGAAAUUUGCUUACGAAUCUCGGCGACAC (((...(((..(((-(...(((.-......))).)))).)))....(((((((((((((((-((..(((-(.(((((........)))))))))..))))))))))))).)))).))).. ( -35.10) >DroSim_CAF1 11522 116 - 1 GUCACGAACUCGUG-CAAAAGUU-AAUAAUACUCGCAUUGUUGAACUCGAAUUUGUGAGCA-AAACGCA-AAGCGUUAAAUGUUAAACGCUUGAAAUUUGCUUACGAAUCUCGGCGACAC ((((((....)))(-(.....((-((((((......))))))))...((((((((((((((-((.....-(((((((........)))))))....))))))))))))).)))))))).. ( -36.00) >DroYak_CAF1 12270 117 - 1 GUCACGAACUCGUG-CAAAAGUU-AAUAAUACUCGCAUUGUUGAACUCGAAUUUGUGAGCA-AAAGGCGAGAGCGUUAAAUGUUAAAUGCUUGAAAUUUGCUUACGAAUCUCGACGACAC ((((((....))).-......((-((((((......))))))))..(((((((((((((((-((......(((((((........)))))))....))))))))))))).)))).))).. ( -32.50) >DroPer_CAF1 11988 106 - 1 GUCACGAGCUCGGGGCCAACGCUGAAUCCCAAUCAUGAUGAU------GGCUUUUCAGGCGCACACACA-AAA-----GGCGAUAAACGAAUGA-AUUUGCGUACCAAUCUCG-CGACAC (((.((((....((.......(((((..(((.((.....)))------))...)))))(((((....((-...-----(........)...)).-...))))).))...))))-.))).. ( -25.80) >consensus GUCACGAACUCGUG_CAAAAGUU_AAUAAUACUCGCAUUGUUGAACUCGAAUUUGUGAGCA_AAACGCA_AAGCGUUAAAUGUUAAACGCUUGAAAUUUGCUUACGAAUCUCGGCGACAC (((.((....))..................................(((((((((((((((.........(((((((........)))))))......))))))))))).)))).))).. (-12.97 = -15.94 + 2.98)
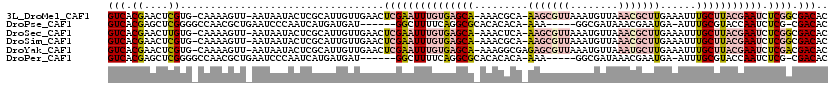
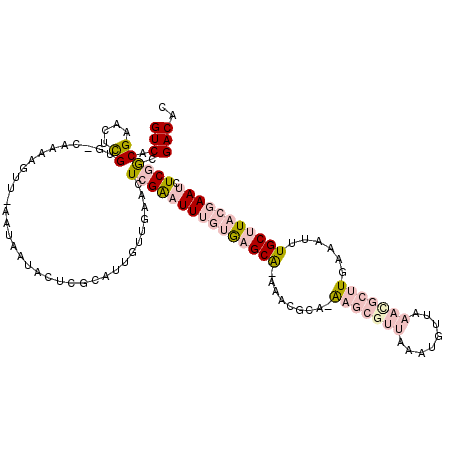
| Location | 1,553,787 – 1,553,878 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 97.80 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -19.81 |
| Energy contribution | -19.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.74 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1553787 91 - 23771897 UGACAAGAGUUUCCCAUUAGGGGAAUAAAAAUCACGUUAAUGAUAACGAAUGAGUCACGAACUCGUGCAAAAGUUAAUAAUACUCGCAUUG ((((....(((((((....))))))).....((((((((....)))))..))))))).......((((...(((.......))).)))).. ( -18.00) >DroSec_CAF1 11499 91 - 1 UGACAAGAGUUUCCCAUUAGGGGAAUAAAAAUCACGUUAGUGAUAACGAAUGAGUCACGAACUUGUGCAAAAGUUAAUAAUACUCGCAUUG ((((....(((((((....))))))).....((((((((....)))))..)))))))..(((((......)))))................ ( -20.40) >DroSim_CAF1 11600 91 - 1 UGACAAGAGUUUCCCAUUAGGGGAAUAAAAAUCACGUUAGUGAUAACGAAUGAGUCACGAACUCGUGCAAAAGUUAAUAAUACUCGCAUUG ((((....(((((((....))))))).....((((((((....)))))..))))))).......((((...(((.......))).)))).. ( -20.30) >DroYak_CAF1 12349 91 - 1 UGACAAGAGUUUCCCAUUAGGGAAAUAAAAACCACGUUAGUGAUAACGAAUGAGUCACGAACUCGUGCAAAAGUUAAUAAUACUCGCAUUG ((((....(((((((....)))))))........(((((....))))).....)))).......((((...(((.......))).)))).. ( -20.60) >consensus UGACAAGAGUUUCCCAUUAGGGGAAUAAAAAUCACGUUAGUGAUAACGAAUGAGUCACGAACUCGUGCAAAAGUUAAUAAUACUCGCAUUG ((((....(((((((....)))))))........(((((....))))).....)))).......((((...(((.......))).)))).. (-19.81 = -19.62 + -0.19)

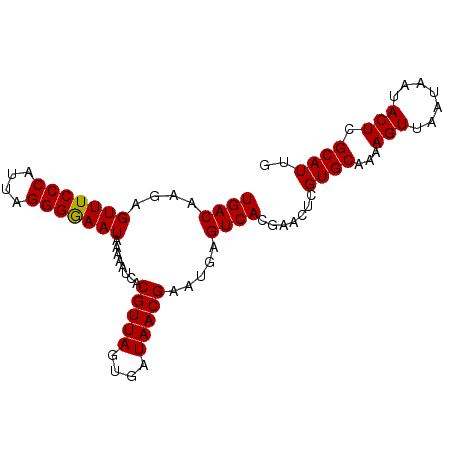
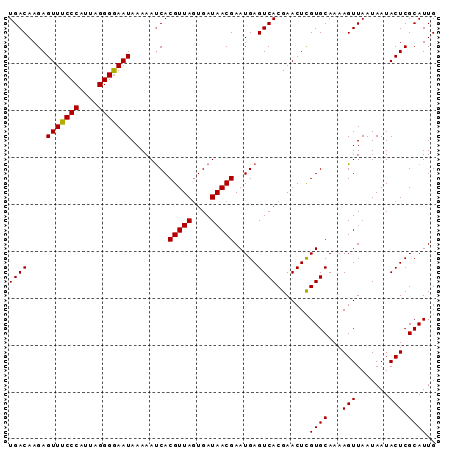
| Location | 1,553,825 – 1,553,918 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -16.33 |
| Consensus MFE | -12.54 |
| Energy contribution | -13.79 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1553825 93 + 23771897 UCAUUCGUUAUCAUUAACGUGAUUUUUAUUCCCCUAAUGGGAAACUCUUGUCAGCACAACUGGAAUACUCACAAUAUACCAUAAGUAUUCAAA .....(((((....)))))((((.....(((((.....)))))......)))).........(((((((..............)))))))... ( -15.74) >DroSec_CAF1 11537 93 + 1 UCAUUCGUUAUCACUAACGUGAUUUUUAUUCCCCUAAUGGGAAACUCUUGUCAGCACAACUGGAAUACUCACAAUUUACAAUAAGUAUUCAAA .........(((((....))))).....(((((.....)))))........(((.....)))(((((((..............)))))))... ( -17.24) >DroSim_CAF1 11638 93 + 1 UCAUUCGUUAUCACUAACGUGAUUUUUAUUCCCCUAAUGGGAAACUCUUGUCAGCACAACUGGAAUACUCACAAUAUACAAUAAGUAUUCAAA .........(((((....))))).....(((((.....)))))........(((.....)))(((((((..............)))))))... ( -17.24) >DroYak_CAF1 12387 89 + 1 UCAUUCGUUAUCACUAACGUGGUUUUUAUUUCCCUAAUGGGAAACUCUUGUCAACACAACUGGAAUACUCACAAUACUC----ACGAUUCAAA ....((((.(((((....))))).....((((((....))))))(..((((....))))..).................----))))...... ( -15.10) >consensus UCAUUCGUUAUCACUAACGUGAUUUUUAUUCCCCUAAUGGGAAACUCUUGUCAGCACAACUGGAAUACUCACAAUAUACAAUAAGUAUUCAAA ......((((((((....))))).....(((((.....))))).........))).......(((((((..............)))))))... (-12.54 = -13.79 + 1.25)

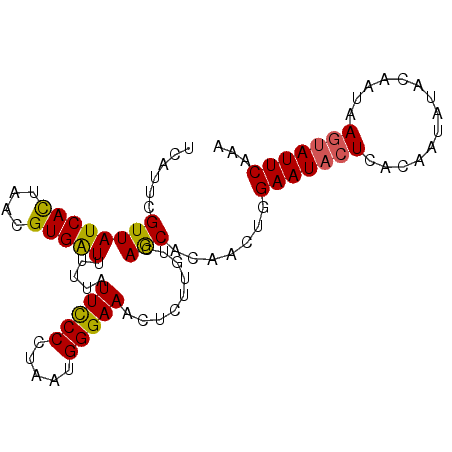
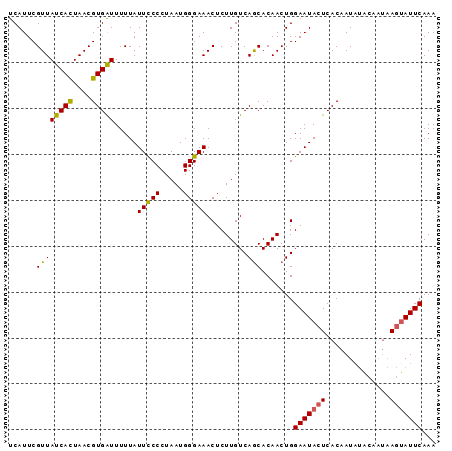
| Location | 1,553,825 – 1,553,918 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 92.65 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1553825 93 - 23771897 UUUGAAUACUUAUGGUAUAUUGUGAGUAUUCCAGUUGUGCUGACAAGAGUUUCCCAUUAGGGGAAUAAAAAUCACGUUAAUGAUAACGAAUGA ...(((((((((..(....)..)))))))))(((.....)))......(((((((....)))))))........(((((....)))))..... ( -23.70) >DroSec_CAF1 11537 93 - 1 UUUGAAUACUUAUUGUAAAUUGUGAGUAUUCCAGUUGUGCUGACAAGAGUUUCCCAUUAGGGGAAUAAAAAUCACGUUAGUGAUAACGAAUGA ...((((((((((........))))))))))..(((((((((((..(((((((((....))))))).....))..)))))).)))))...... ( -26.80) >DroSim_CAF1 11638 93 - 1 UUUGAAUACUUAUUGUAUAUUGUGAGUAUUCCAGUUGUGCUGACAAGAGUUUCCCAUUAGGGGAAUAAAAAUCACGUUAGUGAUAACGAAUGA ...((((((((((........))))))))))..(((((((((((..(((((((((....))))))).....))..)))))).)))))...... ( -26.80) >DroYak_CAF1 12387 89 - 1 UUUGAAUCGU----GAGUAUUGUGAGUAUUCCAGUUGUGUUGACAAGAGUUUCCCAUUAGGGAAAUAAAAACCACGUUAGUGAUAACGAAUGA ......((((----(.((.((((.(((((.......))))).))))..(((((((....)))))))....)))))((((....)))))).... ( -19.10) >consensus UUUGAAUACUUAUUGUAUAUUGUGAGUAUUCCAGUUGUGCUGACAAGAGUUUCCCAUUAGGGGAAUAAAAAUCACGUUAGUGAUAACGAAUGA ...((((((((((........))))))))))(((.....)))......(((((((....)))))))........(((((....)))))..... (-20.70 = -21.57 + 0.87)

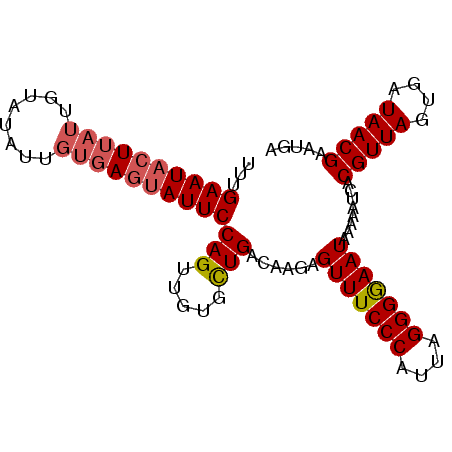
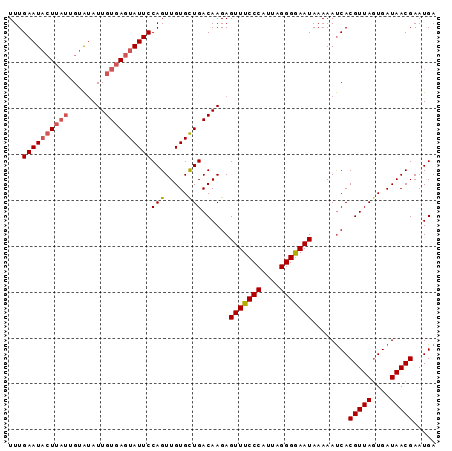
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:26 2006