| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,603,149 – 14,603,245 |
| Length | 96 |
| Max. P | 0.888845 |

| Location | 14,603,149 – 14,603,245 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -15.97 |
| Energy contribution | -17.11 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
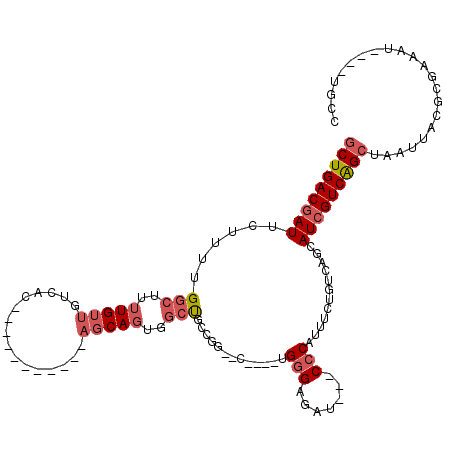
>3L_DroMel_CAF1 14603149 96 - 23771897 GCUGACGAUUCUUUUGGCUUUUGUUGUCAC-----------AGCAGUGGCUGCCGG--C----UGGGAGAU---CCCAUUUCUGUCAGCAUCGUCAGCUAAUUACGCGAAAU----UGCC (((((((((.....((((..(..((((...-----------.))))..)..))))(--(----(((.(((.---......))).)))))))))))))).......(((....----))). ( -36.10) >DroSec_CAF1 2222 96 - 1 GCUGACGAUUCUUUUGGCUUUUGUUGUCAC-----------AGCAGUGGCUGCCGG--C----UGGGAGAU---CCCAUUUCUGUCAGCAUCGUCAGCUAAUUACGCGAAAU----UGCC (((((((((.....((((..(..((((...-----------.))))..)..))))(--(----(((.(((.---......))).)))))))))))))).......(((....----))). ( -36.10) >DroEre_CAF1 5029 106 - 1 GCUGACGAUUCUUUUGGCUUUUGUUGUCACAGUC-----ACAGCAGCGGCUGGCUG--CCGCAUGGGAGAU---CCCAUUUCUGUCAGCAUCGUCAGCUAAUUACGCGAAAU----UGCC (((((((((....(((((...((((((.......-----))))))(((((.....)--))))(((((....---)))))....))))).))))))))).......(((....----))). ( -39.30) >DroYak_CAF1 4915 107 - 1 GCUGACGAUUCUUUUGGCCUUUGUUGUCACAGUCAUCCCACAGCAGCUGCUGCCUA--C----UGGGGGAU---CCCAUUUCUGUCAGCAUCGUCAGCUAAUUACGCGAAAU----UGCC ((((((((.((....))....(((((..((((..(((((.((((....))))((..--.----.)))))))---.......))))))))))))))))).......(((....----))). ( -31.60) >DroAna_CAF1 3752 85 - 1 UCUGACGAUUC----GGCUUUUGUUGUCAC-----------AGCAGG-GCU------------UGGGAGAU---CCCAUUUCUGUCAGCAUCGUCAGCUAAUUACGCGAAAU----UGCC .((((((((.(----(((.......)))..-----------.(((((-(..------------((((....---)))).))))))..).))))))))........(((....----))). ( -25.10) >DroPer_CAF1 7634 98 - 1 GCUGACG----UUUUGCCUUUUGUUGUAGC-----------CUCAUUCCUCGCUCGUUC----CGAGGGAUACACACACUCGUGCGAGU---GUCGGCUAAUUGCGCGAUAUUUAUUCCC .......----..((((.....((..((((-----------(..((((((((.......----))))))))....((((((....))))---)).)))))...))))))........... ( -26.70) >consensus GCUGACGAUUCUUUUGGCUUUUGUUGUCAC___________AGCAGUGGCUGCCGG__C____UGGGAGAU___CCCAUUUCUGUCAGCAUCGUCAGCUAAUUACGCGAAAU____UGCC (((((((((......(((..(((((................)))))..))).............(((.......)))............)))))))))...................... (-15.97 = -17.11 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:26 2006