| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
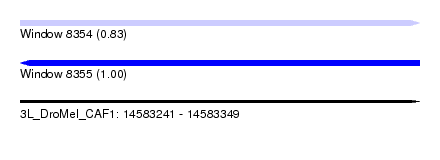
| Location | 14,583,241 – 14,583,349 |
| Length | 108 |
| Max. P | 0.997360 |

| Location | 14,583,241 – 14,583,349 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -31.58 |
| Consensus MFE | -28.05 |
| Energy contribution | -28.24 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
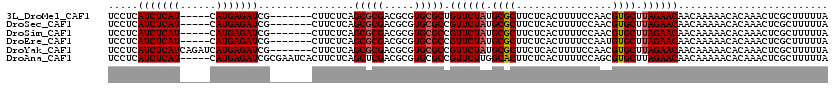
>3L_DroMel_CAF1 14583241 108 + 23771897 UAAAAAGCGAGUUUGUGUUUUUGUUGUUCUAAGCACGUUGGAAAAGUGAGAAGCGCAUAGAACAGCGCACGCGUCGCGCUGAGAAG-------CGAUCUCAUG-----AUGAGAUGAGGA ......((((...(((((...((((((((((.((...((....))((.....)))).)))))))))).)))))))))(((....))-------)..((((((.-----.....)))))). ( -31.20) >DroSec_CAF1 57911 108 + 1 UAAAAAGCGAGUUUGUGUUUUUGUUGUUCUAAGCACGUUGGAAAAGUGAGAAGCGCAUAGAACGGCGCACGCGUCGCGCUGAGAAG-------CGAUCUCAUG-----AUGAGAUGAGGA ......((((...(((((...((((((((((.((...((....))((.....)))).)))))))))).)))))))))(((....))-------)..((((((.-----.....)))))). ( -30.40) >DroSim_CAF1 59887 108 + 1 UAAAAAGCGAGUUUGUGUUUUUGUUGUUCUAAGCACGUUGGAAAAGUGAGAAGCGCAUAGAACGGCGCACGCGUCGCGCUGAGAAG-------CGAUCUCAUG-----AUGAGAUGAGGA ......((((...(((((...((((((((((.((...((....))((.....)))).)))))))))).)))))))))(((....))-------)..((((((.-----.....)))))). ( -30.40) >DroEre_CAF1 57999 108 + 1 UAAAAAGCGAGUUUGUGUUUUUGUUGUUCUAAGCACAUUGGAAAAGUGAGAAGCGCAUAGAACGGCGCACGCGUCGCGCUGAGAAG-------CGAUCUCAUG-----AUGAGAUGAGGA ......((((...((((((((..((.((((((.....)))))).))..))).((((........)))))))))))))(((....))-------)..((((((.-----.....)))))). ( -30.60) >DroYak_CAF1 59790 113 + 1 UAAAAAGCGAGUUUGUGUUUUUGUUGUUCUAAGCACGUUGGAAAAGUGAGAAGCGCAUAGAACGGCGCACGCGUCGCGCUGAGAAG-------CGAUCUCAUGAUCUGAUGAGAUGAGGA ......((((...(((((...((((((((((.((...((....))((.....)))).)))))))))).)))))))))(((....))-------).(((((((......)))))))..... ( -32.40) >DroAna_CAF1 54078 115 + 1 UAAAAAGCGAGUUUGUGUUUUUGUUGUUCUAAGCACGCUGGAAAAGUGAGAAGUGCCAAGAACGGCGCACGCGUCGAGCUGAGAAGUGAUUCGCGAUCUCAUG-----AUGAGAUGAGGA ......(((((((.((((((..........))))))(((.((...(((....(((((......))))).))).)).)))........)))))))..((((((.-----.....)))))). ( -34.50) >consensus UAAAAAGCGAGUUUGUGUUUUUGUUGUUCUAAGCACGUUGGAAAAGUGAGAAGCGCAUAGAACGGCGCACGCGUCGCGCUGAGAAG_______CGAUCUCAUG_____AUGAGAUGAGGA .....((((....(((((...((((((((((.((.(................).)).)))))))))).)))))...))))...............(((((((......)))))))..... (-28.05 = -28.24 + 0.19)

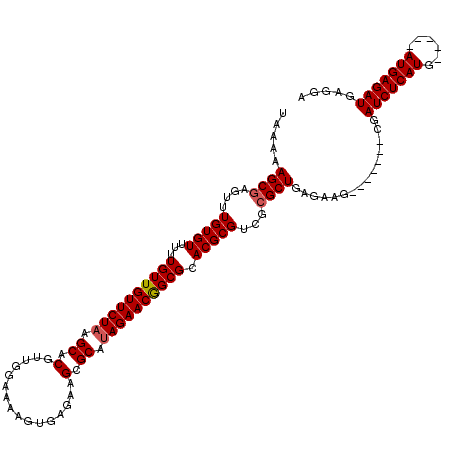
| Location | 14,583,241 – 14,583,349 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.35 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -22.31 |
| Energy contribution | -22.51 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14583241 108 - 23771897 UCCUCAUCUCAU-----CAUGAGAUCG-------CUUCUCAGCGCGACGCGUGCGCUGUUCUAUGCGCUUCUCACUUUUCCAACGUGCUUAGAACAACAAAAACACAAACUCGCUUUUUA .....(((((..-----...))))).(-------(.....((((((.....))))))((((((.((((................)))).)))))).................))...... ( -24.99) >DroSec_CAF1 57911 108 - 1 UCCUCAUCUCAU-----CAUGAGAUCG-------CUUCUCAGCGCGACGCGUGCGCCGUUCUAUGCGCUUCUCACUUUUCCAACGUGCUUAGAACAACAAAAACACAAACUCGCUUUUUA .....(((((..-----...))))).(-------((....)))((((.((....)).((((((.((((................)))).))))))...............))))...... ( -23.69) >DroSim_CAF1 59887 108 - 1 UCCUCAUCUCAU-----CAUGAGAUCG-------CUUCUCAGCGCGACGCGUGCGCCGUUCUAUGCGCUUCUCACUUUUCCAACGUGCUUAGAACAACAAAAACACAAACUCGCUUUUUA .....(((((..-----...))))).(-------((....)))((((.((....)).((((((.((((................)))).))))))...............))))...... ( -23.69) >DroEre_CAF1 57999 108 - 1 UCCUCAUCUCAU-----CAUGAGAUCG-------CUUCUCAGCGCGACGCGUGCGCCGUUCUAUGCGCUUCUCACUUUUCCAAUGUGCUUAGAACAACAAAAACACAAACUCGCUUUUUA .....(((((..-----...))))).(-------((....)))((((.((....)).((((((.((((................)))).))))))...............))))...... ( -24.29) >DroYak_CAF1 59790 113 - 1 UCCUCAUCUCAUCAGAUCAUGAGAUCG-------CUUCUCAGCGCGACGCGUGCGCCGUUCUAUGCGCUUCUCACUUUUCCAACGUGCUUAGAACAACAAAAACACAAACUCGCUUUUUA .....(((((((......))))))).(-------((....)))((((.((....)).((((((.((((................)))).))))))...............))))...... ( -25.39) >DroAna_CAF1 54078 115 - 1 UCCUCAUCUCAU-----CAUGAGAUCGCGAAUCACUUCUCAGCUCGACGCGUGCGCCGUUCUUGGCACUUCUCACUUUUCCAGCGUGCUUAGAACAACAAAAACACAAACUCGCUUUUUA .....(((((..-----...))))).((((...........((.((...)).))...(((((.(((((................))))).)))))...............))))...... ( -23.99) >consensus UCCUCAUCUCAU_____CAUGAGAUCG_______CUUCUCAGCGCGACGCGUGCGCCGUUCUAUGCGCUUCUCACUUUUCCAACGUGCUUAGAACAACAAAAACACAAACUCGCUUUUUA .....(((((((......)))))))................(((((.....))))).((((((.((((................)))).))))))......................... (-22.31 = -22.51 + 0.20)
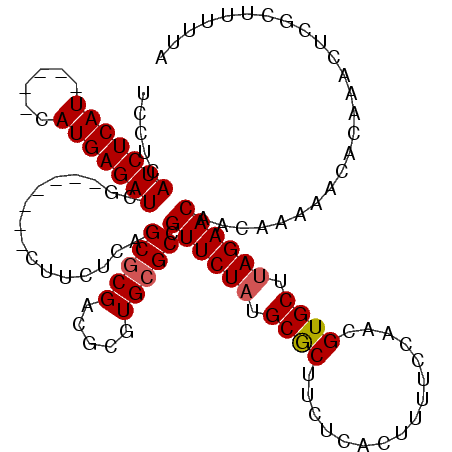
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:19 2006