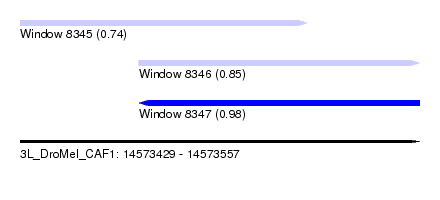
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,573,429 – 14,573,557 |
| Length | 128 |
| Max. P | 0.984136 |

| Location | 14,573,429 – 14,573,521 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 97.10 |
| Mean single sequence MFE | -10.80 |
| Consensus MFE | -10.00 |
| Energy contribution | -10.25 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.743040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14573429 92 + 23771897 AACUAGAUUUCCCAUCCCCUUUCCGCUUGCAGCAUUUUGUCUGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUC ...((((.................((.....))...(((.(((((....)))))))).)))).............................. ( -11.30) >DroSec_CAF1 48288 92 + 1 AACUAGAUUUCCCAUCCCCUUUCCGCUUGCAGCAUUUUGUCUGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUC ...((((.................((.....))...(((.(((((....)))))))).)))).............................. ( -11.30) >DroEre_CAF1 48140 92 + 1 AACUAGAUUUCCCAUCCCCUUUCCGCUUGCAGCAUGUUGUCCGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUC ........................(((.((((.((((.....))))..)))))))..................................... ( -8.40) >DroYak_CAF1 49777 91 + 1 AACUAGAUU-GCCAUCCCCUUUCCGCUUGUAGCAUGUUGUCUGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUC ...((((..-..............((.....))..((((.(((((....))))))))))))).............................. ( -12.20) >consensus AACUAGAUUUCCCAUCCCCUUUCCGCUUGCAGCAUGUUGUCUGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUC ...((((.................((.....))...(((.(((((....)))))))).)))).............................. (-10.00 = -10.25 + 0.25)



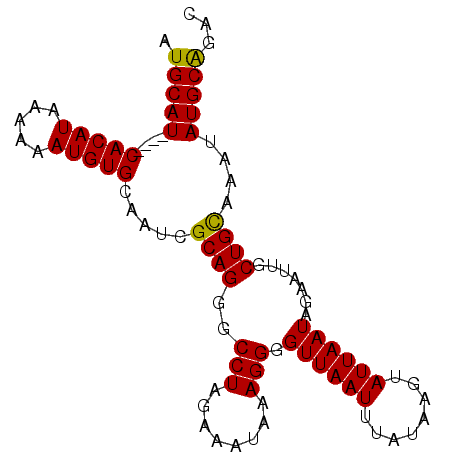
| Location | 14,573,467 – 14,573,557 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 96.51 |
| Mean single sequence MFE | -16.37 |
| Consensus MFE | -13.24 |
| Energy contribution | -13.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14573467 90 + 23771897 GUCUGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUCUAGGCCCUGCGAUUGCACAUUUUUUAUGUG----AUGCAU ...(((((..((((((........(((((........)))))..(((........)))..))))))...(((((.....)))))----))))). ( -16.30) >DroSec_CAF1 48326 90 + 1 GUCUGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUCUAGGCCCUGCGGUUGCACAUUUUUUAUGUG----AUGCAU (.(((((....))))))...........................(((........)))...((((....(((((.....)))))----.)))). ( -15.80) >DroSim_CAF1 50261 90 + 1 GUCUGCAUAUUUACAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUAUCUAGGCCCUGCGAUUGCACAUUUUUUAUGUG----AUGCAU ...(((((.......((((((.......((..((((((........))))))..))........))))))((((.....)))).----))))). ( -13.06) >DroEre_CAF1 48178 90 + 1 GUCCGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUCUAGGCCCUGCGAUUGCACAUUUUUUAUGUG----AUGCAU ....((((..((((((........(((((........)))))..(((........)))..))))))...(((((.....)))))----)))).. ( -16.00) >DroYak_CAF1 49814 94 + 1 GUCUGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUCUAGGCCCUGCGAUUGCACAUUUUUUAUGUGCAUAAUGCAU ...(((((..((((((........(((((........)))))..(((........)))..)))))).(((((((.....)))))))..))))). ( -20.70) >consensus GUCUGCAUAUUUGCAGCAAUUCUAUUAAUACUUAUAAAUUAACCCCUUUAUUUCUAGGCCCUGCGAUUGCACAUUUUUUAUGUG____AUGCAU ....((((..((((((........(((((........)))))..(((........)))..))))))...(((((.....)))))....)))).. (-13.24 = -13.28 + 0.04)

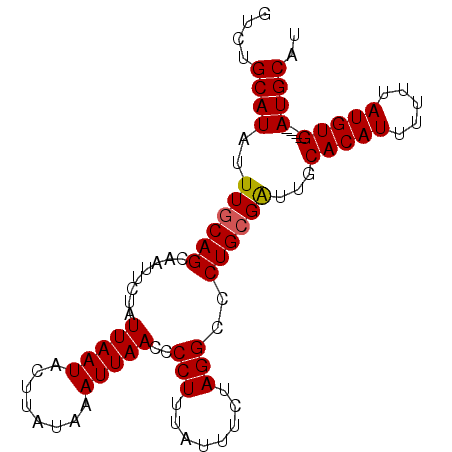
| Location | 14,573,467 – 14,573,557 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 96.51 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -16.66 |
| Energy contribution | -16.34 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14573467 90 - 23771897 AUGCAU----CACAUAAAAAAUGUGCAAUCGCAGGGCCUAGAAAUAAAGGGGUUAAUUUAUAAGUAUUAAUAGAAUUGCUGCAAAUAUGCAGAC .(((((----(((((.....))))).....((((..(((........))).((((((........)))))).......))))....)))))... ( -17.10) >DroSec_CAF1 48326 90 - 1 AUGCAU----CACAUAAAAAAUGUGCAACCGCAGGGCCUAGAAAUAAAGGGGUUAAUUUAUAAGUAUUAAUAGAAUUGCUGCAAAUAUGCAGAC .(((((----(((((.....))))).....((((..(((........))).((((((........)))))).......))))....)))))... ( -17.10) >DroSim_CAF1 50261 90 - 1 AUGCAU----CACAUAAAAAAUGUGCAAUCGCAGGGCCUAGAUAUAAAGGGGUUAAUUUAUAAGUAUUAAUAGAAUUGCUGUAAAUAUGCAGAC .(((((----(((((.....))))).....((((..(((........))).((((((........)))))).......))))....)))))... ( -14.70) >DroEre_CAF1 48178 90 - 1 AUGCAU----CACAUAAAAAAUGUGCAAUCGCAGGGCCUAGAAAUAAAGGGGUUAAUUUAUAAGUAUUAAUAGAAUUGCUGCAAAUAUGCGGAC .(((((----(((((.....))))).....((((..(((........))).((((((........)))))).......))))....)))))... ( -16.40) >DroYak_CAF1 49814 94 - 1 AUGCAUUAUGCACAUAAAAAAUGUGCAAUCGCAGGGCCUAGAAAUAAAGGGGUUAAUUUAUAAGUAUUAAUAGAAUUGCUGCAAAUAUGCAGAC .(((((..(((((((.....)))))))...((((..(((........))).((((((........)))))).......))))....)))))... ( -22.70) >consensus AUGCAU____CACAUAAAAAAUGUGCAAUCGCAGGGCCUAGAAAUAAAGGGGUUAAUUUAUAAGUAUUAAUAGAAUUGCUGCAAAUAUGCAGAC .(((((....(((((.....))))).....((((..(((........))).((((((........)))))).......))))....)))))... (-16.66 = -16.34 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:12 2006