| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,572,794 – 14,572,902 |
| Length | 108 |
| Max. P | 0.971469 |

| Location | 14,572,794 – 14,572,902 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.46 |
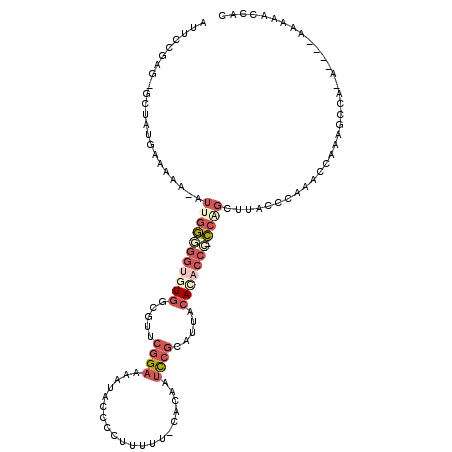
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -8.66 |
| Energy contribution | -10.58 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.32 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
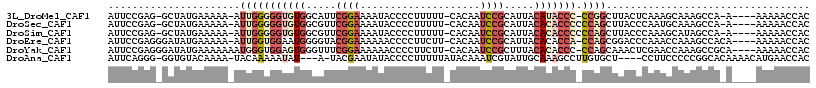
>3L_DroMel_CAF1 14572794 108 - 23771897 AUUCCGAG-GCUAUGAAAAA-AUUGGGGGUGUGGCAUUCGGAAAAUACCCCUUUUU-CACAAUCCGCAUUACAUACCC-CCGGCUUACUCAAAGCAAAGCCA-A----AAAAACCAC .......(-(((........-..((((((((((..((.((((..............-.....)))).))..)))))))-)))((((.....))))..)))).-.----......... ( -31.51) >DroSec_CAF1 47670 109 - 1 AUUCCGAG-GCUAUGAAAAA-AUUGGGGGUGUGGCGUUCGGAAAAUACCCCUUUUU-CACAAUCCGCAUUACACACCCCCCAGCUUACCCAAUGCAAAGCCA-A----AAAAACCAC .......(-(((........-...(((((((((..((.((((..............-.....)))).))..)))))))))..((.........))..)))).-.----......... ( -30.51) >DroSim_CAF1 49607 109 - 1 AUUCCGAG-GCUAUGAAAAA-AUUGGGGGUGUGGCGUUCGGAAAAUACCCCUUUUU-CACAAUCCGCAUUACACACCCCCCAGCUUACCCAAAGCAUAGCCA-A----AAAAACCAC .......(-(((((......-...(((((((((..((.((((..............-.....)))).))..)))))))))..((((.....)))))))))).-.----......... ( -35.21) >DroEre_CAF1 47232 110 - 1 AUUCCGAGGGAUAUGAAAAA-AUUGGUGGAAUGGGGUACGGAAAAAACCCCUUCUU-CACAAUCCGCAUUACACACCA-CCAGCGGACCCAAACCAAAGCCACA----AAAAACCAC .......(((..........-....(((((..(((((.........)))))...))-)))..(((((...........-...))))))))..............----......... ( -24.64) >DroYak_CAF1 49094 111 - 1 AUUCCGAGGGAUAUGAAAAAAAUGGGUGGAGUGGGUUUCGGAAAAAACCCCUUCUU-CACAAUCCGCUUUACACACCC-CCAGCAAACUCGAACCAAAGCCGCA----AAAAACCAC ....((((((((.(....)......((((((.((((((......))))))...)))-))).))))(((..........-..)))...)))).............----......... ( -23.00) >DroAna_CAF1 44547 107 - 1 AUUCAGGG-GGUGUACAAAA-UACAAAAAUAU---A-UACGAAUAUACCCCUUUUUAUACAAAUCGUAUUGCAAAGCCUUGUGCU----CCUUCCCCCGGCACAAAACAUGAACCAC .....(((-((.((((((..-........(((---(-(....)))))...((((..((((.....))))...))))..)))))).----....)))))((..((.....))..)).. ( -20.00) >consensus AUUCCGAG_GCUAUGAAAAA_AUUGGGGGUGUGGCGUUCGGAAAAUACCCCUUUUU_CACAAUCCGCAUUACACACCC_CCAGCUUACCCAAACCAAAGCCA_A____AAAAACCAC ......................(((((((((((.....((((....................)))).....))))))).)))).................................. ( -8.66 = -10.58 + 1.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:10 2006