
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
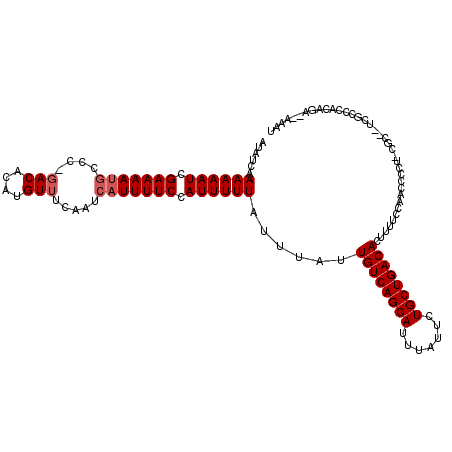
| Location | 14,568,768 – 14,568,881 |
| Length | 113 |
| Max. P | 0.999475 |

| Location | 14,568,768 – 14,568,881 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -19.87 |
| Consensus MFE | -18.45 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14568768 113 + 23771897 AUAUCAAAAAAUCGAAAAUGCCC-GACACAUGUUUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCU-CGC--UCGCCCACAAA--AAAU ......((((((.(((((((...-(((....))).....))))))).))))))......((((((((.......))))))))..............-...--...........--.... ( -19.10) >DroPse_CAF1 45647 118 + 1 AUAUCAAAAAAUCGAAAAUGCCC-GACACAUGUUUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUUUGCUGACACUUUUCUAUCUCCUUUGCUGCCACCCUCCGAUCAGGU ......((((((.(((((((...-(((....))).....))))))).))))))......((((((((.......))))))))........................(((......))). ( -20.90) >DroSec_CAF1 42908 113 + 1 AUAUCAAAAAAUCGAAAACGCCC-GACACAUGU-UCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCUCCGC--UCGCCCACAGA--AAAU ...((.((((((.(((((.(...-(((....))-)....).))))).))))))......((((((((.......))))))))..................--.........))--.... ( -15.70) >DroSim_CAF1 44856 115 + 1 AUAUCAAAAAAUCGAAAAUGCCCGGACACAUGUUUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCUCCGC--UCGCCCACAGA--AAAU ...((.((((((.(((((((...((((....))))....))))))).))))))......((((((((.......))))))))..................--.........))--.... ( -20.50) >DroAna_CAF1 40655 110 + 1 AUAUCAAAAAAUCGAAAAUACCC-GACACAUGUUUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCUGGCACCC-CUC--UU-CC--CAGA--AACC ......((((((.((((((....-(((....)))......)))))).))))))......((((((((.......))))))))..(((((((.....-...--..-.)--))))--)).. ( -22.10) >DroPer_CAF1 48958 115 + 1 AUAUCAAAAAAUCGAAAAUGCCC-GACACAUGUUUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUUUGCUGACACUUUUCUAUCUCCUUUG---CCACCCUCCGAUCAGGU ......((((((.(((((((...-(((....))).....))))))).))))))......((((((((.......)))))))).................---....(((......))). ( -20.90) >consensus AUAUCAAAAAAUCGAAAAUGCCC_GACACAUGUUUCAAUCAUUUUCCAUUUUUAUUUAUUGUCAGCAUUUAUUCUGCUGACACUUUUCCAACCCCU_CGC__UCGCCCACAGA__AAAU ......((((((.(((((((....(((....))).....))))))).))))))......((((((((.......))))))))..................................... (-18.45 = -18.95 + 0.50)

| Location | 14,568,768 – 14,568,881 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.40 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -21.36 |
| Energy contribution | -21.08 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14568768 113 - 23771897 AUUU--UUUGUGGGCGA--GCG-AGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAAACAUGUGUC-GGGCAUUUUCGAUUUUUUGAUAU ..((--(((((......--)))-))))((..(((((.((((((((.......)))))))).............(((((((.((((........))-)).)))))))..)))))..)).. ( -27.60) >DroPse_CAF1 45647 118 - 1 ACCUGAUCGGAGGGUGGCAGCAAAGGAGAUAGAAAAGUGUCAGCAAAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAAACAUGUGUC-GGGCAUUUUCGAUUUUUUGAUAU ((((.......))))......................((((((((.......))))))))......((((((.(((((((.((((........))-)).))))))).))))))...... ( -25.70) >DroSec_CAF1 42908 113 - 1 AUUU--UCUGUGGGCGA--GCGGAGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGA-ACAUGUGUC-GGGCGUUUUCGAUUUUUUGAUAU .(((--(((((......--))))))))((..(((((.((((((((.......)))))))).............(((((((.((((-.(....)))-)).)))))))..)))))..)).. ( -30.40) >DroSim_CAF1 44856 115 - 1 AUUU--UCUGUGGGCGA--GCGGAGGGGUUGGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAAACAUGUGUCCGGGCAUUUUCGAUUUUUUGAUAU .(((--(((((......--))))))))((..(((((.((((((((.......)))))))).............(((((((.(((..((....)).))).)))))))..)))))..)).. ( -29.10) >DroAna_CAF1 40655 110 - 1 GGUU--UCUG--GG-AA--GAG-GGGUGCCAGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAAACAUGUGUC-GGGUAUUUUCGAUUUUUUGAUAU ..((--((((--(.-(.--...-...).)))))))..((((((((.......))))))))......((((((.(((((((.((((........))-)).))))))).))))))...... ( -29.70) >DroPer_CAF1 48958 115 - 1 ACCUGAUCGGAGGGUGG---CAAAGGAGAUAGAAAAGUGUCAGCAAAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAAACAUGUGUC-GGGCAUUUUCGAUUUUUUGAUAU ((((.......))))..---.................((((((((.......))))))))......((((((.(((((((.((((........))-)).))))))).))))))...... ( -25.70) >consensus AUUU__UCUGUGGGCGA__GCG_AGGGGUUAGAAAAGUGUCAGCAGAAUAAAUGCUGACAAUAAAUAAAAAUGGAAAAUGAUUGAAACAUGUGUC_GGGCAUUUUCGAUUUUUUGAUAU .....................................((((((((.......))))))))......((((((.(((((((......((....)).....))))))).))))))...... (-21.36 = -21.08 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:07 2006