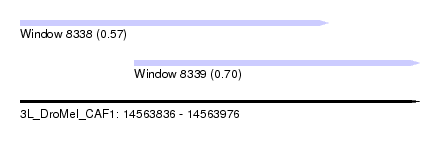
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,563,836 – 14,563,976 |
| Length | 140 |
| Max. P | 0.704734 |

| Location | 14,563,836 – 14,563,944 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -54.97 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
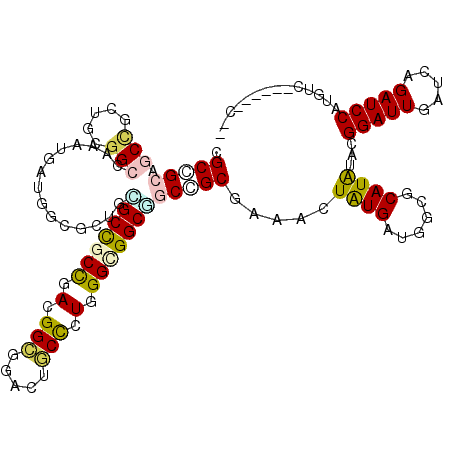
>3L_DroMel_CAF1 14563836 108 + 23771897 UCGCAUGGGCAUGGACAUCCGCGAAGCCAUUGCUCAUGGCCGCCG------------CCAAAUGAUGACGUUGAGCCGCCGACGGGGGACUUCCCUGGGCGGCGGCCGCAAAACUAUGAU ..((((((((((((.(.((...)).)))).)))))))((((((((------------((.........(((((......)))))((((....)))).))))))))))))........... ( -51.50) >DroVir_CAF1 43525 117 + 1 UCGCAUGUGCAUGAACAUCGGCAAAUCCAUUGCUCAGACCUGCGGCAGCCGCUGCGGCCAAAUGAUGGCG---UGCCGCCGACGGCGGACUGCUCUGGGAGGCAGCCGCAAAACUGUGAU (((((..(((..(.....)(((...(((...((........))(((((((((((((((...(((....))---)...)))).)))))).)))))...)))....))))))....))))). ( -44.40) >DroGri_CAF1 44633 120 + 1 UCGCAUGCGCAUGCACAUCAGCGAAACCAUUGCUCAAGCCCGCUGCAGCUGCAGCGGCCAAAUGGUGACGCUGUGCCGCCGACGGCGGACUGCCCUGGGCGGCGGCAGCGAAACUAUGAU ((((.(((..........(((((..((((((((....))(((((((....)))))))...))))))..)))))(((((((.(.(((.....))).).))))))))))))))......... ( -58.20) >DroWil_CAF1 187444 120 + 1 UGGCAUGAGCAUGGACAUCGGCGAAUCCGUUGCUCAGGCCAGCCGCUGCCGCUGCAGCAAGAUGGUGUCUCUGCGCUGCUGACGGCGGACUUCCCUGGGUGGCGGCGGCGAAGGUAUGGU ((((.((((((((((..........)))).)))))).))))(((((((((((((((((.(((.......)))..)))))...(((.((....)))))))))))))))))........... ( -59.30) >DroMoj_CAF1 49654 120 + 1 UCGCAUGUGCAUGAACAUCGGCGAAUCCAUUGCUCAGUCCCGCGGCAGCCGCUGCGGCCAGGUGAUGGCGCUGCGCCGCCGACGGCGGGCUGCUCUGGGAGGCAGCGGCGAAGCUGUGAU ((((((((......)))).(((....((.(((((...(((((.(((((((((.(((.(((.....)))))).))((((....)))).))))))).)))))))))).))....))))))). ( -55.90) >DroAna_CAF1 35889 108 + 1 UCGCAUGGGCGUGGACAUCCGCGAAUCCAUUGCUCAUGGCCGCCG------------CCAAAUGGUGGCGCUGGGCCGCCGACGGGGGCGAUCCCUGGGCGGCCGCCGCGAAGCUGUGAU ..((((((.(((((....)))))...))).)))(((((((.((((------------((....))))))((.((((((((..(((((.....)))))))))))).).))...))))))). ( -60.50) >consensus UCGCAUGGGCAUGGACAUCGGCGAAUCCAUUGCUCAGGCCCGCCGCAGCCGCUGCGGCCAAAUGAUGGCGCUGCGCCGCCGACGGCGGACUGCCCUGGGCGGCGGCCGCGAAACUAUGAU ..((....((.....((((.((((.....))))........((.((.......)).)).....))))......(((((((.(.(((.....))).).))))))))).))........... (-24.58 = -24.58 + 0.00)

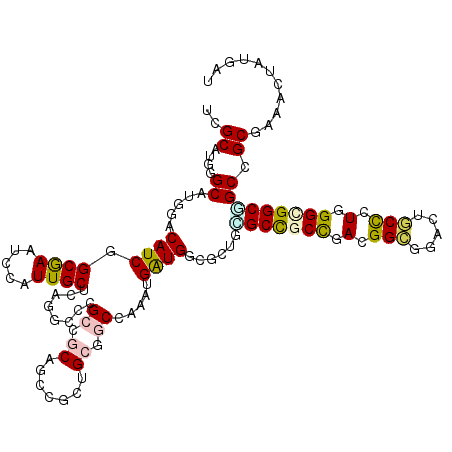
| Location | 14,563,876 – 14,563,976 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.36 |
| Mean single sequence MFE | -48.63 |
| Consensus MFE | -25.75 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14563876 100 + 23771897 CGCCG------------CCAAAUGAUGACGUUGAGCCGCCGACGGGGGACUUCCCUGGGCGGCGGCCGCAAAACUAUGAUGGCGCAUAUACGGAUUUAUCAGAUCCAUCUC------G-- .((.(------------(((..(.(((...(((.(((((((.(.((((....)))).).)))))))..)))...))).)))))))......(((((.....))))).....------.-- ( -39.70) >DroVir_CAF1 43565 109 + 1 UGCGGCAGCCGCUGCGGCCAAAUGAUGGCG---UGCCGCCGACGGCGGACUGCUCUGGGAGGCAGCCGCAAAACUGUGAUGGCGCAUAUAGGGAUUGAUGAGAUCCAUGUC------C-- .(((((....))))).((((.....))))(---(((((.((...((((.(((((......))))))))).......)).))))))......(((((.....))))).....------.-- ( -42.60) >DroGri_CAF1 44673 114 + 1 CGCUGCAGCUGCAGCGGCCAAAUGGUGACGCUGUGCCGCCGACGGCGGACUGCCCUGGGCGGCGGCAGCGAAACUAUGAUGCCGCAUAUAGGGAUUGAUGAGAUCCAUCUC------CGU .(((((....)))))(((...(((((..((((((((((((.(.(((.....))).).)))))).))))))..)))))...)))........(((..((((.....))))))------).. ( -55.20) >DroWil_CAF1 187484 112 + 1 AGCCGCUGCCGCUGCAGCAAGAUGGUGUCUCUGCGCUGCUGACGGCGGACUUCCCUGGGUGGCGGCGGCGAAGGUAUGGUGUCUCAUAUACGGAUUUAUCAGAUCCAUUUC------U-- .(((((((((((((((((.(((.......)))..)))))...(((.((....)))))))))))))))))((((((((((....))))))..(((((.....))))).))))------.-- ( -50.10) >DroMoj_CAF1 49694 112 + 1 CGCGGCAGCCGCUGCGGCCAGGUGAUGGCGCUGCGCCGCCGACGGCGGGCUGCUCUGGGAGGCAGCGGCGAAGCUGUGAUGGCGCAUGUAGGGAUUUAUUAGAUCCAUGUC------G-- ((((((.((((((((.((((.....))))((.((.(((((...))))))).))........))))))))...))))))..((((.......(((((.....))))).))))------.-- ( -52.90) >DroAna_CAF1 35929 106 + 1 CGCCG------------CCAAAUGGUGGCGCUGGGCCGCCGACGGGGGCGAUCCCUGGGCGGCCGCCGCGAAGCUGUGAUGCCGCAUAUACGGAUUGAUCAGAUCCAUGUCCACCGGC-- (((((------------((....)))))))...(((((((..(((((.....))))))))))))((((((..((......)))))......(((((.....))))).........)))-- ( -51.30) >consensus CGCCGCAGCCGCUGCGGCCAAAUGAUGGCGCUGCGCCGCCGACGGCGGACUGCCCUGGGCGGCGGCCGCGAAACUAUGAUGGCGCAUAUACGGAUUGAUCAGAUCCAUGUC______C__ .(((((.(((.....)))...............(((((((.(.(((.....))).).)))))))))))).....((((......))))...(((((.....))))).............. (-25.75 = -26.28 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:05 2006