| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,549,237 – 14,549,387 |
| Length | 150 |
| Max. P | 0.851460 |

| Location | 14,549,237 – 14,549,357 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.94 |
| Mean single sequence MFE | -43.97 |
| Consensus MFE | -38.55 |
| Energy contribution | -38.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14549237 120 - 23771897 GGUCUGGUUCCAGAAUCGUCGCACCAAAUGGCGCAAACGGCAUGCGGCGGAAAUGGCCACGGCCAAAAGGAAACAGGACGAUAUGGGCGGCGAUAACGAUGGCGAUUGCAGCGAGACCAU (((((.((((((..((((((((.((....)).))....(((.((.(((.......))).)))))....(....)..)))))).)))...(((((..(....)..)))))))).))))).. ( -44.90) >DroVir_CAF1 27231 120 - 1 GGUUUGGUUUCAGAAUCGUCGCACCAAAUGGCGCAAACGUCAUGCGGCGGAAAUGGCCACGGCCAAGCGGAAACAGGACGACAUGGGCGGCGACAACGAUGGAGACUGCAGCGAGACCAU (((((.(((.(((..(((((((.(((.((((((....)))))).((((.....((((....)))).))(....)....))...))))))))))((....))....))).))).))))).. ( -43.60) >DroGri_CAF1 27612 120 - 1 GGUUUGGUUUCAGAAUCGUCGCACCAAAUGGCGCAAACGUCAUGCGGCGGAAAUGGCAACGGCCAAGCGGAAACAGGAUGACAUGGGUGGCGACAACGAUGGCGACUGCAGCGAGACCAU (((((.(((.(((..((((((((.....(((((....)))))))))))))...((((....)))).(((....).......(((.(.((....)).).)))))..))).))).))))).. ( -43.00) >DroWil_CAF1 164250 120 - 1 GGUUUGGUUUCAGAAUCGUCGCACCAAAUGGCGCAAACGUCAUGCGGCCGAAAUGGCCACAGCGAAGCGGAAACAGGACGACAUGGGUGGCGAUAACGAUGGCGAUUGCAGCGAGACCAU (((((.(((.((..((((((((.((....)).))...((((((.(((((.....))))..........(....)..........).))))))........)))))))).))).))))).. ( -41.60) >DroMoj_CAF1 30297 120 - 1 GGUUUGGUUUCAGAAUCGUCGCACCAAAUGGCGCAAGCGUCAUGCGGCGGAAAUGGCAACGGCCAAGCGGAAACAGGACGACAUGGGUGGCGACAACGACGGCGACUGCAGCGAGACCAU (((((.(((.(((....((((((((.....((....))..((((((((.....((((....)))).))(....)....)).))))))).)))))..((....)).))).))).))))).. ( -44.80) >DroAna_CAF1 22759 120 - 1 GGUCUGGUUCCAGAACCGUCGCACCAAAUGGCGCAAACGGCAUGCGGCGGAAAUGGCCACGGCCAAGAGGAAACAGGACGACAUGGGCGGCGACAACGACGGGGACUGCAGCGAGGCCAU (((((.(((.(((..(((((((.(((..(((((((.......)))(((.......)))...))))...(....).........)))))(....)...)))))...))).))).))))).. ( -45.90) >consensus GGUUUGGUUUCAGAAUCGUCGCACCAAAUGGCGCAAACGUCAUGCGGCGGAAAUGGCCACGGCCAAGCGGAAACAGGACGACAUGGGCGGCGACAACGAUGGCGACUGCAGCGAGACCAU (((((.(((.(((..(((((((.(((...(.((((.......)))).).....((((....))))...(....).........))))))))))...(......).))).))).))))).. (-38.55 = -38.05 + -0.50)

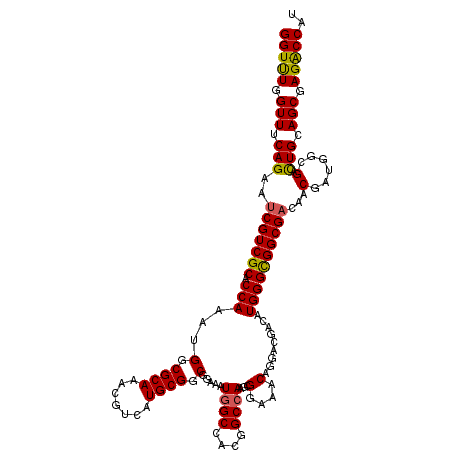
| Location | 14,549,277 – 14,549,387 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -22.08 |
| Energy contribution | -21.88 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14549277 110 + 23771897 CGUCCUGUUUCCUUUUGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAGACCUGCAACAAAA-------CCAAGAAAUAAC--UA-CGCUUU (((...(((...((((((..(((((.......))))).(((((((.(((((....)))))))))..((((....))))...)))......-------))))))...)))--.)-)).... ( -32.90) >DroPse_CAF1 25444 102 + 1 CGUCCUGUUUCCUCUUGGCCGUGGCCAUUUCCGCCGCAUGCCGCUUGCGCCAUUUGGUGCGGCGAUUCUGGAACCAAACCUGCA----------------AAACGUAAG--UAAAACUUU ..(((.(..((.....(((.(((((.......)))))..)))(((.(((((....))))))))))..).)))............----------------..((....)--)........ ( -29.70) >DroGri_CAF1 27652 112 + 1 CAUCCUGUUUCCGCUUGGCCGUUGCCAUUUCCGCCGCAUGACGUUUGCGCCAUUUGGUGCGACGAUUCUGAAACCAAACCUGUAACAAUU-------CCAAAUUGAAAUGGCA-AACUUU .....................((((((((((..........((((.(((((....)))))))))....((....))..............-------.......)))))))))-)..... ( -24.10) >DroWil_CAF1 164290 105 + 1 CGUCCUGUUUCCGCUUCGCUGUGGCCAUUUCGGCCGCAUGACGUUUGCGCCAUUUGGUGCGACGAUUCUGAAACCAAACCUGUAAAAAAG-------GGGAAACAA--------AACUUU .((..(((((((...(((.(((((((.....))))))))))((((.(((((....)))))))))..............(((.......))-------)))))))).--------.))... ( -38.10) >DroAna_CAF1 22799 100 + 1 CGUCCUGUUUCCUCUUGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGGUUCUGGAACCAGACCUGCAAAAAAA-------CCAACAAGGA------------- ..(((((((......((((....))))........(((.((((((.(((((....)))))))))))((((....))))..))).......-------..))).))))------------- ( -33.70) >DroPer_CAF1 28992 118 + 1 CGUCCUGUUUCCUCUUGGCCGUGGCCAUUUCCGCCGCAUGCCGCUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAAACCUGCAACGGAAAAUGUAAGCGAAACGUAAG--UAAAACUUU ......(((((((..((((....))))((((((..(((...((.(((((((....)))))))))....((....))....)))..)))))).....)).))))).....--......... ( -33.00) >consensus CGUCCUGUUUCCUCUUGGCCGUGGCCAUUUCCGCCGCAUGCCGUUUGCGCCAUUUGGUGCGACGAUUCUGGAACCAAACCUGCAACAAAA_______CCAAAACGUAA___UA_AACUUU ......(((((.........(((((.......)))))....((.(((((((....))))))))).....))))).............................................. (-22.08 = -21.88 + -0.19)

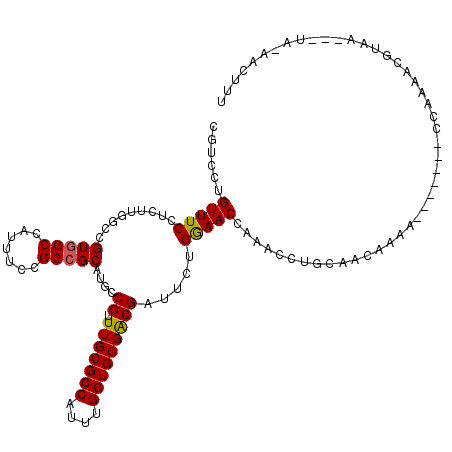
| Location | 14,549,277 – 14,549,387 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.18 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -26.88 |
| Energy contribution | -26.35 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
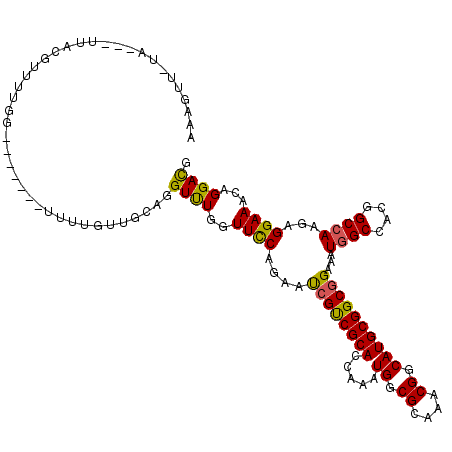
>3L_DroMel_CAF1 14549277 110 - 23771897 AAAGCG-UA--GUUAUUUCUUGG-------UUUUGUUGCAGGUCUGGUUCCAGAAUCGUCGCACCAAAUGGCGCAAACGGCAUGCGGCGGAAAUGGCCACGGCCAAAAGGAAACAGGACG ......-..--(((.((((((..-------((((((((((..((((....)))).((((.((.((....)).))..))))..)))))))))).((((....))))..))))))...))). ( -33.40) >DroPse_CAF1 25444 102 - 1 AAAGUUUUA--CUUACGUUU----------------UGCAGGUUUGGUUCCAGAAUCGCCGCACCAAAUGGCGCAAGCGGCAUGCGGCGGAAAUGGCCACGGCCAAGAGGAAACAGGACG ...((...(--(....))..----------------.))..((((..((((....((((((((((.....((....))))..))))))))...((((....))))...))))...)))). ( -33.30) >DroGri_CAF1 27652 112 - 1 AAAGUU-UGCCAUUUCAAUUUGG-------AAUUGUUACAGGUUUGGUUUCAGAAUCGUCGCACCAAAUGGCGCAAACGUCAUGCGGCGGAAAUGGCAACGGCCAAGCGGAAACAGGAUG ...(((-.((((((((..(((((-------(((((.........))))))))))..(((((((.....(((((....)))))))))))))))))))))))........(....)...... ( -35.60) >DroWil_CAF1 164290 105 - 1 AAAGUU--------UUGUUUCCC-------CUUUUUUACAGGUUUGGUUUCAGAAUCGUCGCACCAAAUGGCGCAAACGUCAUGCGGCCGAAAUGGCCACAGCGAAGCGGAAACAGGACG ...(((--------((((((((.-------((........(((((.......))))).((((.....((((((....))))))(.((((.....)))).).)))))).))))))))))). ( -36.30) >DroAna_CAF1 22799 100 - 1 -------------UCCUUGUUGG-------UUUUUUUGCAGGUCUGGUUCCAGAACCGUCGCACCAAAUGGCGCAAACGGCAUGCGGCGGAAAUGGCCACGGCCAAGAGGAAACAGGACG -------------.....(((.(-------(((((((...(((((((..(((...((((((((((....)).((.....)).))))))))...)))))).))))..))))))))..))). ( -36.30) >DroPer_CAF1 28992 118 - 1 AAAGUUUUA--CUUACGUUUCGCUUACAUUUUCCGUUGCAGGUUUGGUUCCAGAAUCGUCGCACCAAAUGGCGCAAGCGGCAUGCGGCGGAAAUGGCCACGGCCAAGAGGAAACAGGACG .........--.....(((((.(((....(((((((((((.(((((((....((....))..))))))).((....))....)))))))))))((((....)))).))))))))...... ( -40.60) >consensus AAAGUU_UA___UUACGUUUUGG_______UUUUGUUGCAGGUUUGGUUCCAGAAUCGUCGCACCAAAUGGCGCAAACGGCAUGCGGCGGAAAUGGCCACGGCCAAGAGGAAACAGGACG .........................................((((..((((....((((((((.....((.((....)).))))))))))...((((....))))...))))...)))). (-26.88 = -26.35 + -0.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:46:00 2006