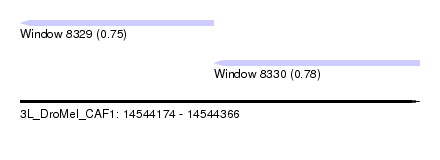
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,544,174 – 14,544,366 |
| Length | 192 |
| Max. P | 0.780786 |

| Location | 14,544,174 – 14,544,267 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -11.65 |
| Energy contribution | -11.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14544174 93 - 23771897 CAAGCGAUGGAGUCCAUUUUGAGUUUAUUGUUUGAUGAUUUGAACAUUUUAUA----UGUGUACGUUUG----------GCUCUGUGUGUGUGUGUGUGUUAGUCGC (((((((((((.((......)).)))))))))))..(((...(((((..((((----((..((((....----------....))))..)))))).))))).))).. ( -24.30) >DroSec_CAF1 18217 89 - 1 CAAGCGAUGGAGUCCAUUUUGAGUUUAUUGUUUGAUGAUUUGAACAUUUUAUA----UGUGUAUGUUUG----------GCUCUGUGUG----UGUGAGUUAGUCGC (((((((((((.((......)).)))))))))))..((((.((((((......----.....))))))(----------((((......----...))))))))).. ( -19.00) >DroWil_CAF1 152773 89 - 1 CAAGCGAUGGACUCCAUUUUGAGUUUAUUGUUUGAUGAUUUGAACAUUUUUUA----UAUGCAUAUCUC----------AGCAAGUAUG----UGAGAGGUCGUUGG ((((((((((((((......))))))))))))))........(((((((((((----(((((.......----------.....)))))----)))))))).))).. ( -26.40) >DroYak_CAF1 19838 89 - 1 CAAGCGAUGGAGUCCAUUUUGAGUUUAUUGUUUGAUGAUUUGAACAUUUUAUA----UGUGUAUGUUUG----------CCUCUGCGUG----UGUGUGUUAGUCGC (((((((((((.((......)).)))))))))))..(((...(((((..((((----((((((....))----------)....)))))----)).))))).))).. ( -22.00) >DroAna_CAF1 17674 99 - 1 CAAGCGAUGGAGUCCAUUUUGAGUUUAUUGUUUGAUGAUUUGAACAUUUUAUA----UAUGUAUGUGUGUGCGCCUGUAUCUCUGCAUG----UGUGAGUUAGUCCU (((((((((((.((......)).))))))))))).......((....((((((----((((((.(.(.((((....))))).)))))))----))))))....)).. ( -24.40) >DroPer_CAF1 23351 81 - 1 CAAGCGAUGGAGUCCAUUUUGAGUUUAUUGUUUGAUGAUUUGAACAUUUUAUAUACAUGUGUAUGUAUA----------GCCAUGUGUG----U------------G (((((((((((.((......)).)))))))))))...............((((((((((.((.......----------))))))))))----)------------) ( -21.60) >consensus CAAGCGAUGGAGUCCAUUUUGAGUUUAUUGUUUGAUGAUUUGAACAUUUUAUA____UGUGUAUGUUUG__________GCUCUGUGUG____UGUGAGUUAGUCGC (((((((((((.((......)).)))))))))))......................................................................... (-11.65 = -11.65 + 0.00)


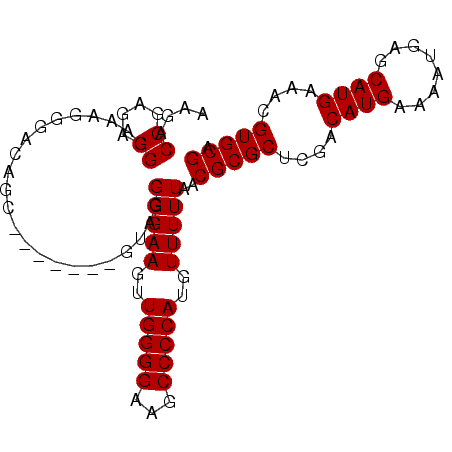
| Location | 14,544,267 – 14,544,366 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.81 |
| Mean single sequence MFE | -26.84 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.22 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14544267 99 - 23771897 AAGACCGAGAGGGAAAGGGACAGCAUAGACAGUAGGGGAAGUUGGGGAAGCCCCAUGUUUUUAACGCGCUCGACAUGAAAAUGAGCAUGAAACGUGCGA ....((....))....(((...((..(((((...((((............)))).))))).....)).)))..(((....))).(((((...))))).. ( -23.40) >DroSec_CAF1 18306 92 - 1 AAGACCGAGCGGGAAAGGGACAGC-------GUAGGGGAAGUUGGGGAAGCCCCAUGUUUUUAACGCGCUCGACAUGAAAAUGAGCAUGAAACGUGCGA .....(((((.(........).((-------((..((((...(((((...)))))..))))..))))))))).(((....))).(((((...))))).. ( -29.80) >DroSim_CAF1 18804 92 - 1 AAGACCGAGCGGGAAAGGGACAGC-------GUAGGGGAAGUUGGGGAAGCCCCAUGUUUUUAACGCGCUCGACAUGAAAAUGAGCAUGAAACGUGCGA .....(((((.(........).((-------((..((((...(((((...)))))..))))..))))))))).(((....))).(((((...))))).. ( -29.80) >DroEre_CAF1 18748 92 - 1 CAGACCGAGAGGGAAAGGGACAGC-------GAAGGGGAAGUUGGGGAAGCCCCAUGUUUUUAACGCGCUCGACAUGAAAAUGAGCAUGAAACGUGCGA ....((....))....(((...((-------(...(((((..(((((...)))))..)))))..))).)))..(((....))).(((((...))))).. ( -26.70) >DroAna_CAF1 17773 75 - 1 -----------------AGACAGC-------GAGGGGGAAGGUGGGGAUGCCCCAUGUUUUUAACGCGCUCGACAUGAAAAUGAGCAUGAAACGUGCGA -----------------....(((-------(...(((((.((((((...)))))).)))))....))))...(((....))).(((((...))))).. ( -24.50) >consensus AAGACCGAGAGGGAAAGGGACAGC_______GUAGGGGAAGUUGGGGAAGCCCCAUGUUUUUAACGCGCUCGACAUGAAAAUGAGCAUGAAACGUGCGA ....((....)).......................(((((..(((((...)))))..)))))..(((((....((((........))))....))))). (-19.12 = -19.22 + 0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:56 2006