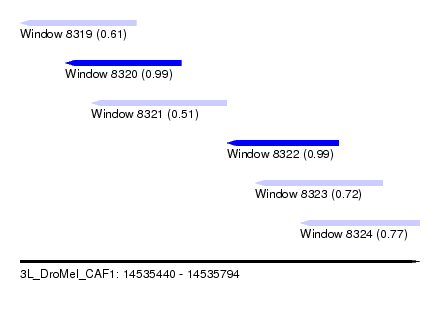
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 14,535,440 – 14,535,794 |
| Length | 354 |
| Max. P | 0.992174 |

| Location | 14,535,440 – 14,535,543 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -34.74 |
| Consensus MFE | -22.01 |
| Energy contribution | -23.10 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14535440 103 - 23771897 GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCUCACAGGCCGCGUCUAAAAAUGCCC--CGCAAGUCAUAGGCAGUUGGAUAUACUCUUUCCUCUGGCAGGGUA ((((((((((...))))))))))((((....(((....)))...((((((...((((.--(....).....)))).)))))).............))))...... ( -33.50) >DroSec_CAF1 9764 103 - 1 GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCC--UGCAAGUCAUAGGCAGCUGGAUAUACUCUUUCCUUUGGCAGGGUG ((((((((((...)))))))))).......(((((.((((..((((......))))))--))...((((.(((.((..(......)..)).))).)))).))))) ( -36.90) >DroSim_CAF1 10172 103 - 1 GACACGUGAUCCGGUCGCGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCA--UGCAAGUCAUAGGCAGCUGGAUAUACUCUUUCCUAUGGCAGAGUA ((((((((((...))))))))))((((....(((....)))...(((((....(((((--((.....))).))))..))))).............))))...... ( -33.80) >DroEre_CAF1 9905 103 - 1 GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCGC--UGCAAGUCACAGACGGCUGUGUAUACCCUUUCUUCUUGCAGGGUA ((((((((((...))))))))))(((.....(((....)))(((((......)))))(--((((((.(((((....))))).............)))))))))). ( -39.14) >DroYak_CAF1 10659 103 - 1 GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAUAUUUCC--UGCAAGUCACAGCCAGCUGGGAAUACCCUUGCUUCUGUCAGAGUA ((((((((((...))))))))))....((((....(((((..(((......(((((((--.((..((....))..)).)))))))....))).)))))..)))). ( -31.00) >DroAna_CAF1 8851 99 - 1 GACACGUGAUCCGGUCACGUGUCACCAAUUCGCUUGCAGGCCACAGCCAAAAAUGGUCAAGGGAA---AUAGCAGACUUGGUAUACUCUUUAAAC---GAGAGGA ((((((((((...))))))))))(((((.(((((......((...((((....))))....))..---..))).)).)))))...(((((.....---))))).. ( -34.10) >consensus GACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCC__UGCAAGUCAUAGGCAGCUGGAUAUACUCUUUCCUCUGGCAGAGUA ((((((((((...)))))))))).(((....(((....)))...(((((....(((.....))).....)))))...)))...((((((.((....)).)))))) (-22.01 = -23.10 + 1.09)


| Location | 14,535,480 – 14,535,583 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -36.56 |
| Energy contribution | -35.95 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14535480 103 - 23771897 CGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCUCACAGGCCGCGUCUAAAAAUGCCC--CGCAA .((((......(.((((((.(((((((......)))))))((((((((((...))))))))))..)))))).))))).((..((((......)))).)--).... ( -36.20) >DroSec_CAF1 9804 103 - 1 CGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCC--UGCAA ((((....)))).((((((.(((((((......)))))))((((((((((...))))))))))..)))))).....((((..((((......))))))--))... ( -38.60) >DroSim_CAF1 10212 103 - 1 CGUGAUUACAUGAGAGUUGACACGCUGCCUGGUUGGUGUGGACACGUGAUCCGGUCGCGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCA--UGCAA ........((((.(.(((.(.((((.(((((...((((.(((((((((((...)))))))))).).....))))..))))).)))).)...))).)))--))... ( -36.80) >DroEre_CAF1 9945 103 - 1 CGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCGC--UGCAA ((((....)))).((((((.(((((((......)))))))((((((((((...))))))))))..))))))(((....))).((((........))))--..... ( -39.40) >DroYak_CAF1 10699 103 - 1 CGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAUAUUUCC--UGCAA ((((.........((((((.(((((((......)))))))((((((((((...))))))))))..))))))(((....))))))).............--..... ( -37.40) >DroAna_CAF1 8885 105 - 1 CGUGAUUACAAGAGAGUUGACACGCCGCCUGGUCGGUGUGGACACGUGAUCCGGUCACGUGUCACCAAUUCGCUUGCAGGCCACAGCCAAAAAUGGUCAAGGGAA ..((((((((((.((((((.(((((((......)))))))((((((((((...))))))))))..)))))).))))..(((....))).....))))))...... ( -43.60) >consensus CGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGGCCGCGUCUAAAAAUGCCC__UGCAA ((((.........((((((.(((((((......)))))))((((((((((...))))))))))..))))))(((....))))))).................... (-36.56 = -35.95 + -0.61)


| Location | 14,535,503 – 14,535,623 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.78 |
| Mean single sequence MFE | -44.30 |
| Consensus MFE | -41.34 |
| Energy contribution | -41.20 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14535503 120 - 23771897 UAUCCGAUGUCGUCGGACAGUCCCAAGGCCACAUGUUUGCCGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCUCACAGG ..((((((...)))))).....((..(((.........)))((((......(.((((((.(((((((......)))))))((((((((((...))))))))))..)))))).))))).)) ( -42.40) >DroSec_CAF1 9827 120 - 1 UAUCCGAUGUCGUCGGACAGUCCCAAGGCCACAUGUUUGCCGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGG ..((((((...)))))).........(((..(((((...........))))).((((((.(((((((......)))))))((((((((((...))))))))))..)))))))))...... ( -44.00) >DroSim_CAF1 10235 120 - 1 UAUCCGAUGUCGUCGGACAGUCCCAAGGCCACAUGUUUGCCGUGAUUACAUGAGAGUUGACACGCUGCCUGGUUGGUGUGGACACGUGAUCCGGUCGCGUGUCGCCAAUUCGCCCACAGG ..((((((...)))))).........(((..(((((...........))))).((((((.((((((........))))))((((((((((...))))))))))..)))))))))...... ( -41.30) >DroEre_CAF1 9968 120 - 1 UAUCCGAUGUCGUCGGACAGUCCCAAGGCCACAUGUUUGCCGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGG ..((((((...)))))).........(((..(((((...........))))).((((((.(((((((......)))))))((((((((((...))))))))))..)))))))))...... ( -44.00) >DroYak_CAF1 10722 120 - 1 UAUCCGAUGUCGUCGGACAGUCCCAAGGCCACAUGUUUGCCGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGG ..((((((...)))))).........(((..(((((...........))))).((((((.(((((((......)))))))((((((((((...))))))))))..)))))))))...... ( -44.00) >DroAna_CAF1 8910 120 - 1 AAGCCGAUGUCGGCGGACAGUCCCAAGGCCACAUGUUUGCCGUGAUUACAAGAGAGUUGACACGCCGCCUGGUCGGUGUGGACACGUGAUCCGGUCACGUGUCACCAAUUCGCUUGCAGG ...(((((..((((((((((((....)))....)))))))))..))).((((.((((((.(((((((......)))))))((((((((((...))))))))))..)))))).))))..)) ( -50.10) >consensus UAUCCGAUGUCGUCGGACAGUCCCAAGGCCACAUGUUUGCCGUGAUUACAUGAGAGUUGACACGCCGCCUGGUUGGUGUGGACACGUGAUCCGGUCACGUGUCGCCAAUUCGCCCACAGG ..((((((...)))))).........(((..(((((...........))))).((((((.(((((((......)))))))((((((((((...))))))))))..)))))))))...... (-41.34 = -41.20 + -0.14)


| Location | 14,535,623 – 14,535,722 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -21.66 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14535623 99 - 23771897 A------CCGACCACCGCCCAAUGCCCAACCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCAAUUUCAUUGGGCAUCAUAUCA-----GAUAUCGGUCGUA .------.(((((...((((((((.........((.((((((..((((............))))..)))))).))))))))))(((......-----)))...))))).. ( -30.10) >DroSec_CAF1 9947 78 - 1 A------C---------------------CCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCAAUUUCAUUGGGCAUCAUAUCA-----GAUAUCGGUCGGA .------(---------------------(.((.....(((....)))............(((((((((.....))))))))).........-----......))..)). ( -16.30) >DroSim_CAF1 10355 78 - 1 A------C---------------------CCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCAAUUUCAUUGGGCAUCAUAUCA-----GAUAUCGGUCGGA .------(---------------------(.((.....(((....)))............(((((((((.....))))))))).........-----......))..)). ( -16.30) >DroEre_CAF1 10088 84 - 1 --------------------------CACCCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCCAUUUCAUUGGGCAUCAUAUCACCGGAUAUAUCGGUCGGA --------------------------.....((....((....))((((...........(((((((((.....)))))))))...(((((....)))))...)))))). ( -18.20) >DroYak_CAF1 10842 105 - 1 ACGCCCACCACCCACCGCCCAAUGCCCACCCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCCAUUUCAUUGGGCAUCACAUCA-----GAUAUCGGUCGGA ..........((.(((((((((((..............(((....)))...(((((((....)))))))......))))))))(((......-----)))...))).)). ( -27.40) >consensus A______C__________________CA_CCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCAAUUUCAUUGGGCAUCAUAUCA_____GAUAUCGGUCGGA .............................................((((...........(((((((((.....)))))))))...(((((......))))).))))... (-15.56 = -15.96 + 0.40)



| Location | 14,535,648 – 14,535,761 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -18.63 |
| Consensus MFE | -13.72 |
| Energy contribution | -13.08 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14535648 113 - 23771897 CGCGGCAAACCGCAAAAACAACGCCCC-UCGGCCACGCCCACCGACCACCGCCCAAUGCCCAACCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCAAUUUCAUUG .(.((((....((.........(((..-..)))..((.....))......))....))))).........((((((..((((............))))..))))))........ ( -19.80) >DroSec_CAF1 9972 91 - 1 CGCGGCAAACCGCAA-AACAACGCCCC-UCGGCCACGCCCAC---------------------CCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCAAUUUCAUUG .((((....))))..-......(((..-..))).........---------------------.......((((((..((((............))))..))))))........ ( -19.20) >DroSim_CAF1 10380 92 - 1 CGCGGCAAACCGCAAAAACAACGCCCC-UCGGCCACGCCCAC---------------------CCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCAAUUUCAUUG .((((....)))).........(((..-..))).........---------------------.......((((((..((((............))))..))))))........ ( -19.20) >DroEre_CAF1 10118 92 - 1 CGCGGCAAACCGCAAAAACAACGCCCC-UCCACCACGCC---------------------CACCCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCCAUUUCAUUG .((((....))))..............-...........---------------------...................(((((....((((....)))))))))......... ( -14.00) >DroAna_CAF1 9065 96 - 1 CGCGGCAAACCGCAAAAGCACCGCCCCCUCAGUACCGCCCUC----------------UCCUCCUCCAACCCUCGG--GUGGCAACUAUUAAACGCUUAAUGGCAAUUUCAUUG .((((....)))).........(((.....(((.((((((..----------------................))--))))..)))(((((....)))))))).......... ( -20.97) >consensus CGCGGCAAACCGCAAAAACAACGCCCC_UCGGCCACGCCCAC__________________C__CCCCAACUUGCCAACAGGCCAACUAUUAAACGCUUAAUGGCAAUUUCAUUG .((((....)))).........(((...............................................(((....))).....(((((....)))))))).......... (-13.72 = -13.08 + -0.64)



| Location | 14,535,688 – 14,535,794 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -20.90 |
| Consensus MFE | -14.84 |
| Energy contribution | -15.88 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 14535688 106 - 23771897 ACCAUUCUACCAUUGAAUGAUGAGGCGACGGCACGCGGCAAACCGCAAAAACAACGCCCC-UCGGCCACGCCCACCGACCACCGCCCAAUGCCCAACCCCAACUUGC ..(((((.......))))).((.((((..(((..((((....)))).........(((..-..))).................)))...))))))............ ( -25.10) >DroSec_CAF1 10012 84 - 1 ACCAUUAUACCAAUGAAUGAUGAGGCGACGGCACGCGGCAAACCGCAA-AACAACGCCCC-UCGGCCACGCCCAC---------------------CCCCAACUUGC ..((((((........)))))).((((..(((..((((....))))..-.....((....-.))))).))))...---------------------........... ( -20.30) >DroSim_CAF1 10420 85 - 1 ACCAUUAUACCAAUGAAUGAUGAGGCGACGGCACGCGGCAAACCGCAAAAACAACGCCCC-UCGGCCACGCCCAC---------------------CCCCAACUUGC ..((((((........)))))).((((..(((..((((....))))........((....-.))))).))))...---------------------........... ( -20.30) >DroEre_CAF1 10158 85 - 1 ACCAUUAUGCCAAUGAAUGAUGAGGCGCCGGCACGCGGCAAACCGCAAAAACAACGCCCC-UCCACCACGCC---------------------CACCCCCAACUUGC ..((((((........)))))).((((..((...((((....)))).........(....-..).)).))))---------------------.............. ( -15.70) >DroAna_CAF1 9103 86 - 1 --CAUU---CACAUGAAUGAUGAGGCGACGGCACGCGGCAAACCGCAAAAGCACCGCCCCCUCAGUACCGCCCUC----------------UCCUCCUCCAACCCUC --((((---(....)))))..((((.((.(((..((((....))))....((...))............))).))----------------.))))........... ( -23.10) >consensus ACCAUUAUACCAAUGAAUGAUGAGGCGACGGCACGCGGCAAACCGCAAAAACAACGCCCC_UCGGCCACGCCCAC__________________C__CCCCAACUUGC ..((((((........)))))).((((.......((((....)))).........(((.....)))..))))................................... (-14.84 = -15.88 + 1.04)

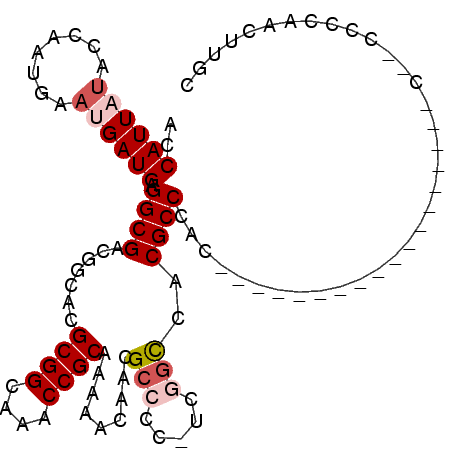
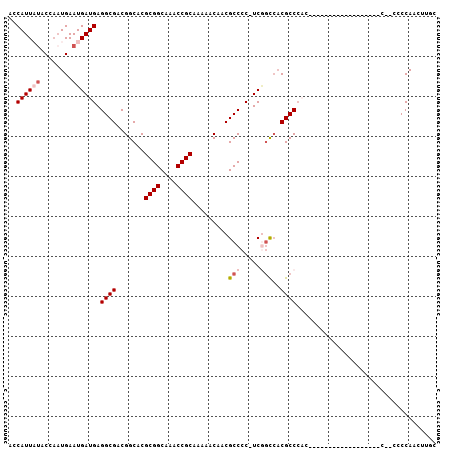
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:45:51 2006